The Pseudorabies Virus DNA Polymerase Accessory Subunit UL42 Directs Nuclear Transport of the Holoenzyme
- PMID: 26913023
- PMCID: PMC4753316
- DOI: 10.3389/fmicb.2016.00124
The Pseudorabies Virus DNA Polymerase Accessory Subunit UL42 Directs Nuclear Transport of the Holoenzyme
Abstract
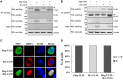
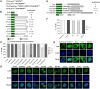
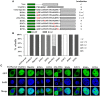
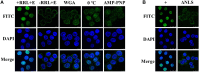
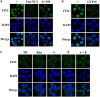
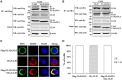
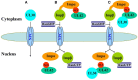
Pseudorabies virus (PRV) DNA replication occurs in the nuclei of infected cells and requires the viral DNA polymerase. The PRV DNA polymerase comprises a catalytic subunit, UL30, and an accessory subunit, UL42, that confers processivity to the enzyme. Its nuclear localization is a prerequisite for its enzymatic function in the initiation of viral DNA replication. However, the mechanisms by which the PRV DNA polymerase holoenzyme enters the nucleus have not been determined. In this study, we characterized the nuclear import pathways of the PRV DNA polymerase catalytic and accessory subunits. Immunofluorescence analysis showed that UL42 localizes independently in the nucleus, whereas UL30 alone predominantly localizes in the cytoplasm. Intriguingly, the localization of UL30 was completely shifted to the nucleus when it was coexpressed with UL42, demonstrating that nuclear transport of UL30 occurs in an UL42-dependent manner. Deletion analysis and site-directed mutagenesis of the two proteins showed that UL42 contains a functional and transferable bipartite nuclear localization signal (NLS) at amino acids 354-370 and that K(354), R(355), and K(367) are important for the NLS function, whereas UL30 has no NLS. Coimmunoprecipitation assays verified that UL42 interacts with importins α3 and α4 through its NLS. In vitro nuclear import assays demonstrated that nuclear accumulation of UL42 is a temperature- and energy-dependent process and requires both importins α and β, confirming that UL42 utilizes the importin α/β-mediated pathway for nuclear entry. In an UL42 NLS-null mutant, the UL42/UL30 heterodimer was completely confined to the cytoplasm when UL42 was coexpressed with UL30, indicating that UL30 utilizes the NLS function of UL42 for its translocation into the nucleus. Collectively, these findings suggest that UL42 contains an importin α/β-mediated bipartite NLS that transports the viral DNA polymerase holoenzyme into the nucleus in an in vitro expression system.
Keywords: DNA polymerase; UL42; accessory subunit; nuclear transport; pseudorabies virus.
Figures

Similar articles
-
Specific inhibition of the interaction between pseudorabies virus DNA polymerase subunits UL30 and UL42 by a synthetic peptide.Vet Microbiol. 2022 Sep;272:109517. doi: 10.1016/j.vetmic.2022.109517. Epub 2022 Jul 20. Vet Microbiol. 2022. PMID: 35908441
-
Nuclear import of HSV-1 DNA polymerase processivity factor UL42 is mediated by a C-terminally located bipartite nuclear localization signal.Biochemistry. 2008 Dec 30;47(52):13764-77. doi: 10.1021/bi800869y. Biochemistry. 2008. PMID: 19053255
-
Ivermectin inhibits DNA polymerase UL42 of pseudorabies virus entrance into the nucleus and proliferation of the virus in vitro and vivo.Antiviral Res. 2018 Nov;159:55-62. doi: 10.1016/j.antiviral.2018.09.010. Epub 2018 Sep 26. Antiviral Res. 2018. PMID: 30266338
-
Targeting the pseudorabies virus DNA polymerase processivity factor UL42 by RNA interference efficiently inhibits viral replication.Antiviral Res. 2016 Aug;132:219-24. doi: 10.1016/j.antiviral.2016.06.010. Epub 2016 Jul 4. Antiviral Res. 2016. PMID: 27387827
-
Nucleocytoplasmic protein transport and recycling of Ran.Cell Struct Funct. 1999 Dec;24(6):425-33. doi: 10.1247/csf.24.425. Cell Struct Funct. 1999. PMID: 10698256 Review.
Cited by
-
Nucleocytoplasmic Shuttling of Porcine Parvovirus NS1 Protein Mediated by the CRM1 Nuclear Export Pathway and the Importin α/β Nuclear Import Pathway.J Virol. 2022 Jan 12;96(1):e0148121. doi: 10.1128/JVI.01481-21. Epub 2021 Oct 13. J Virol. 2022. PMID: 34643426 Free PMC article.
-
Terminase Large Subunit Provides a New Drug Target for Herpesvirus Treatment.Viruses. 2019 Mar 5;11(3):219. doi: 10.3390/v11030219. Viruses. 2019. PMID: 30841485 Free PMC article. Review.
-
Progress of Research into Novel Drugs and Potential Drug Targets against Porcine Pseudorabies Virus.Viruses. 2022 Aug 11;14(8):1753. doi: 10.3390/v14081753. Viruses. 2022. PMID: 36016377 Free PMC article. Review.
-
HSV-1 DNA Replication-Coordinated Regulation by Viral and Cellular Factors.Viruses. 2021 Oct 7;13(10):2015. doi: 10.3390/v13102015. Viruses. 2021. PMID: 34696446 Free PMC article. Review.
-
DNA from Dust: Comparative Genomics of Large DNA Viruses in Field Surveillance Samples.mSphere. 2016 Oct 5;1(5):e00132-16. doi: 10.1128/mSphere.00132-16. eCollection 2016 Sep-Oct. mSphere. 2016. PMID: 27747299 Free PMC article.
References
-
- Allen T. D., Cronshaw J. M., Bagley S., Kiseleva E., Goldberg M. W. (2000). The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113, 1651–1659. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

