Cis regulatory motifs and antisense transcriptional control in the apicomplexan Theileria parva
- PMID: 26896950
- PMCID: PMC4761415
- DOI: 10.1186/s12864-016-2444-5
Cis regulatory motifs and antisense transcriptional control in the apicomplexan Theileria parva
Abstract
Background: Theileria parva is an intracellular parasite that causes a lymphoproliferative disease in cattle. It does so by inducing cancer-like phenotypes in the host cells it infects, although the molecular and regulatory mechanisms involved remain poorly understood. RNAseq data, and the resulting updated genome annotation now available for this parasite, offer an unprecedented opportunity to characterize the genomic features associated with gene regulation in this species. Our previous analyses revealed a T. parva genome even more gene-dense than previously thought, with many adjacent loci overlapping each other, not only at the level of untranslated sequences (UTRs) but even in coding sequences.
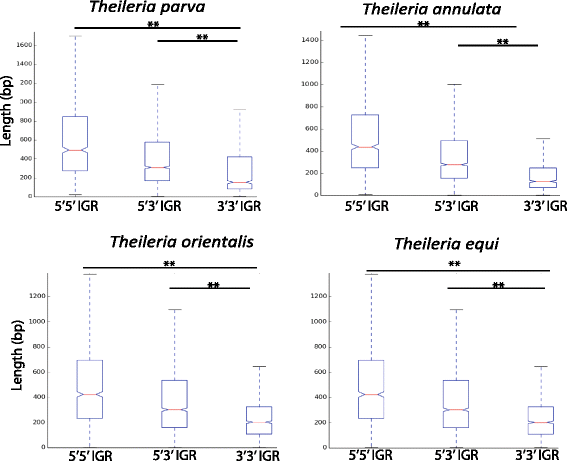
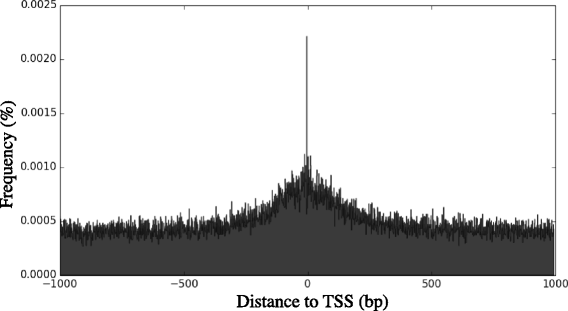
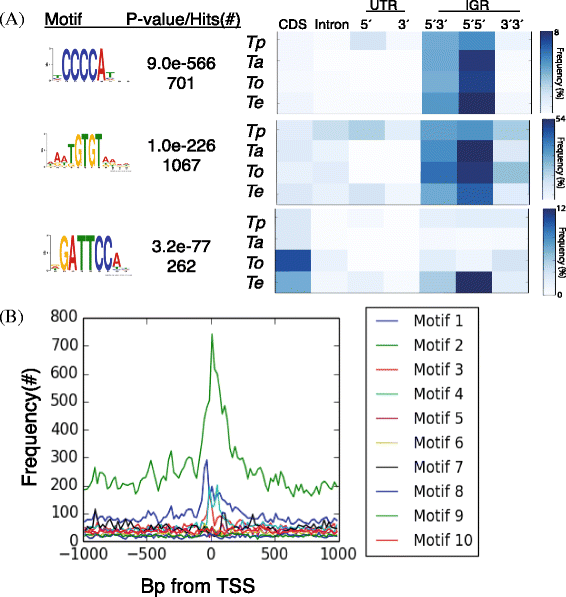
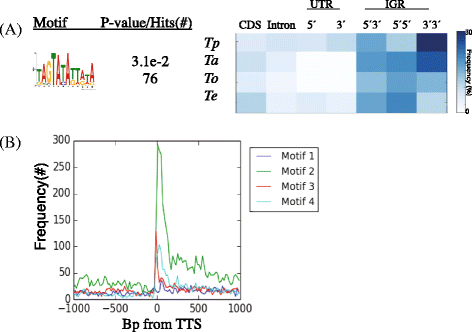
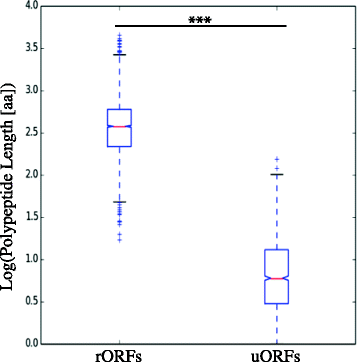
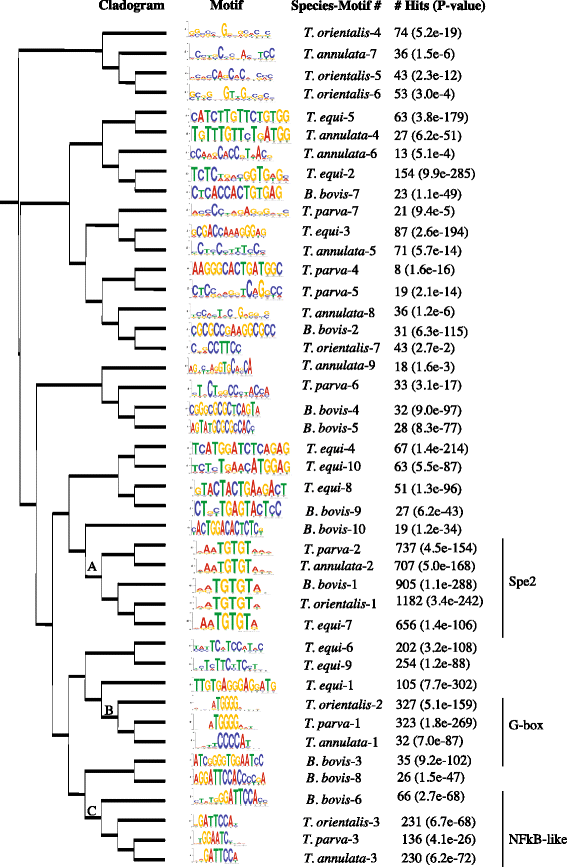
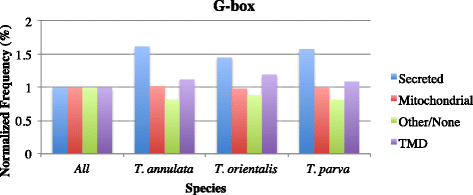
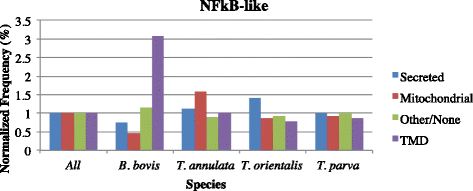
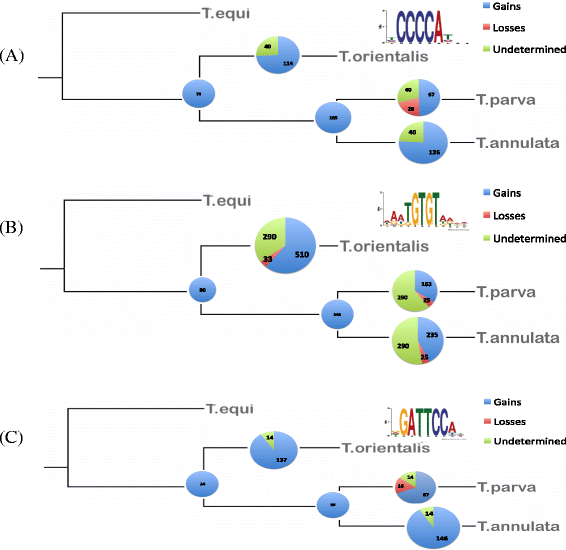
Results: Despite this compactness, Theileria intergenic regions show a pattern of size distribution indicative of monocistronic gene transcription. Three previously described motifs are conserved among Theileria species and highly prevalent in promoter regions near or at the transcription start sites. We found novel motifs at many transcription termination sites, as well as upstream of parasite genes thought to be critical for host transformation. Adjacent genes that could be regulated by antisense transcription from an overlapping transcriptional unit are syntenic between T. parva and P. falciparum at a frequency higher than expected by chance, suggesting the presence of common, and evolutionary old, regulatory mechanisms in the phylum Apicomplexa.
Conclusions: We propose a model of transcription with conserved sense and antisense transcription from a few taxonomically ubiquitous and several species-specific promoter motifs. Interestingly, the gene networks regulated by conserved promoters are themselves, in most cases, not conserved between species or genera.
Figures
Similar articles
-
Re-annotation of the Theileria parva genome refines 53% of the proteome and uncovers essential components of N-glycosylation, a conserved pathway in many organisms.BMC Genomics. 2020 Apr 3;21(1):279. doi: 10.1186/s12864-020-6683-0. BMC Genomics. 2020. PMID: 32245418 Free PMC article.
-
Properties of non-coding DNA and identification of putative cis-regulatory elements in Theileria parva.BMC Genomics. 2008 Dec 3;9:582. doi: 10.1186/1471-2164-9-582. BMC Genomics. 2008. PMID: 19055776 Free PMC article.
-
Microsequence heterogeneity and expression of the LSU rRNA genes within the two single copy ribosomal transcription units of Theileria parva.Gene. 2000 Oct 31;257(2):299-305. doi: 10.1016/s0378-1119(00)00386-3. Gene. 2000. PMID: 11080596
-
Highly syntenic and yet divergent: a tale of two Theilerias.Infect Genet Evol. 2009 Jul;9(4):453-61. doi: 10.1016/j.meegid.2009.01.002. Epub 2009 Jan 19. Infect Genet Evol. 2009. PMID: 19460310 Review.
-
Theileria parva genomics reveals an atypical apicomplexan genome.Int J Parasitol. 2000 Apr 10;30(4):465-74. doi: 10.1016/s0020-7519(00)00016-3. Int J Parasitol. 2000. PMID: 10731569 Review.
Cited by
-
Annotated draft genome sequences of three species of Cryptosporidium: Cryptosporidium meleagridis isolate UKMEL1, C. baileyi isolate TAMU-09Q1 and C. hominis isolates TU502_2012 and UKH1.Pathog Dis. 2016 Oct;74(7):ftw080. doi: 10.1093/femspd/ftw080. Epub 2016 Aug 12. Pathog Dis. 2016. PMID: 27519257 Free PMC article.
-
Approaches to vaccination against Theileria parva and Theileria annulata.Parasite Immunol. 2016 Dec;38(12):724-734. doi: 10.1111/pim.12388. Parasite Immunol. 2016. PMID: 27647496 Free PMC article. Review.
-
Antibodies to in silico selected GPI-anchored Theileria parva proteins neutralize sporozoite infection in vitro.Vet Immunol Immunopathol. 2018 May;199:8-14. doi: 10.1016/j.vetimm.2018.03.004. Epub 2018 Mar 12. Vet Immunol Immunopathol. 2018. PMID: 29678234 Free PMC article.
-
Comparative Transcriptomics of the Bovine Apicomplexan Parasite Theileria parva Developmental Stages Reveals Massive Gene Expression Variation and Potential Vaccine Antigens.Front Vet Sci. 2020 Jun 9;7:287. doi: 10.3389/fvets.2020.00287. eCollection 2020. Front Vet Sci. 2020. PMID: 32582776 Free PMC article.
-
Re-annotation of the Theileria parva genome refines 53% of the proteome and uncovers essential components of N-glycosylation, a conserved pathway in many organisms.BMC Genomics. 2020 Apr 3;21(1):279. doi: 10.1186/s12864-020-6683-0. BMC Genomics. 2020. PMID: 32245418 Free PMC article.
References
-
- Vollmer D. Enhancing the Effectiveness of Sustainability Partnerships Summary of a Workshop. 500 Fifth Street, N.W., Washington DC 20001: The National Academies Press; 2009.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

