Exposure of phosphatidylserine on the cell surface
- PMID: 26891692
- PMCID: PMC4987739
- DOI: 10.1038/cdd.2016.7
Exposure of phosphatidylserine on the cell surface
Abstract
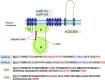
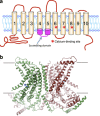
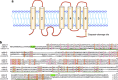
Phosphatidylserine (PtdSer) is a phospholipid that is abundant in eukaryotic plasma membranes. An ATP-dependent enzyme called flippase normally keeps PtdSer inside the cell, but PtdSer is exposed by the action of scramblase on the cell's surface in biological processes such as apoptosis and platelet activation. Once exposed to the cell surface, PtdSer acts as an 'eat me' signal on dead cells, and creates a scaffold for blood-clotting factors on activated platelets. The molecular identities of the flippase and scramblase that work at plasma membranes have long eluded researchers. Indeed, their identity as well as the mechanism of the PtdSer exposure to the cell surface has only recently been revealed. Here, we describe how PtdSer is exposed in apoptotic cells and in activated platelets, and discuss PtdSer exposure in other biological processes.
Figures


Similar articles
-
Flippase and scramblase for phosphatidylserine exposure.Curr Opin Immunol. 2020 Feb;62:31-38. doi: 10.1016/j.coi.2019.11.009. Epub 2019 Dec 11. Curr Opin Immunol. 2020. PMID: 31837595 Review.
-
An Apoptotic 'Eat Me' Signal: Phosphatidylserine Exposure.Trends Cell Biol. 2015 Nov;25(11):639-650. doi: 10.1016/j.tcb.2015.08.003. Epub 2015 Oct 1. Trends Cell Biol. 2015. PMID: 26437594 Review.
-
Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure.Science. 2014 Jun 6;344(6188):1164-8. doi: 10.1126/science.1252809. Science. 2014. PMID: 24904167
-
Phospholipid scrambling on the plasma membrane.Methods Enzymol. 2014;544:381-93. doi: 10.1016/B978-0-12-417158-9.00015-7. Methods Enzymol. 2014. PMID: 24974298
-
Phosphorylation-mediated activation of mouse Xkr8 scramblase for phosphatidylserine exposure.Proc Natl Acad Sci U S A. 2019 Feb 19;116(8):2907-2912. doi: 10.1073/pnas.1820499116. Epub 2019 Feb 4. Proc Natl Acad Sci U S A. 2019. PMID: 30718401 Free PMC article.
Cited by
-
Itaconate impairs immune control of Plasmodium by enhancing mtDNA-mediated PD-L1 expression in monocyte-derived dendritic cells.Cell Metab. 2024 Mar 5;36(3):484-497.e6. doi: 10.1016/j.cmet.2024.01.008. Epub 2024 Feb 6. Cell Metab. 2024. PMID: 38325373 Free PMC article.
-
Arginine-Rich Cell-Penetrating Peptides Induce Lipid Rearrangements for Their Active Translocation across Laterally Heterogeneous Membranes.Adv Sci (Weinh). 2024 Aug;11(32):e2404563. doi: 10.1002/advs.202404563. Epub 2024 Jun 26. Adv Sci (Weinh). 2024. PMID: 38932459 Free PMC article.
-
Multifaceted Roles of ALG-2 in Ca(2+)-Regulated Membrane Trafficking.Int J Mol Sci. 2016 Aug 26;17(9):1401. doi: 10.3390/ijms17091401. Int J Mol Sci. 2016. PMID: 27571067 Free PMC article. Review.
-
Roles of phosphatidylserine exposed on the viral envelope and cell membrane in HIV-1 replication.Cell Commun Signal. 2019 Oct 21;17(1):132. doi: 10.1186/s12964-019-0452-1. Cell Commun Signal. 2019. PMID: 31638994 Free PMC article. Review.
-
Phospholipid flippases enable precursor B cells to flee engulfment by macrophages.Proc Natl Acad Sci U S A. 2018 Nov 27;115(48):12212-12217. doi: 10.1073/pnas.1814323115. Epub 2018 Oct 24. Proc Natl Acad Sci U S A. 2018. PMID: 30355768 Free PMC article.
References
-
- Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys 2010; 39: 407–427. - PubMed
-
- Balasubramanian K, Schroit AJ. Aminophospholipid asymmetry: a matter of life and death. Annu Rev Physiol 2003; 65: 701–734. - PubMed
-
- Zachowski A, Henry JP, Devaux PF. Control of transmembrane lipid asymmetry in chromaffin granules by an ATP-dependent protein. Nature 1989; 340: 75–76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

