Phosphorylation of Bovine Herpesvirus 1 VP8 Plays a Role in Viral DNA Encapsidation and Is Essential for Its Cytoplasmic Localization and Optimal Virion Incorporation
- PMID: 26889039
- PMCID: PMC4836321
- DOI: 10.1128/JVI.00219-16
Phosphorylation of Bovine Herpesvirus 1 VP8 Plays a Role in Viral DNA Encapsidation and Is Essential for Its Cytoplasmic Localization and Optimal Virion Incorporation
Abstract
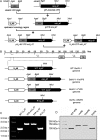
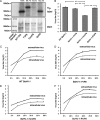
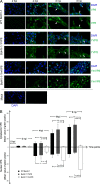
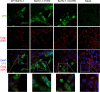
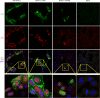
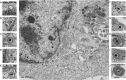
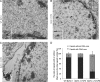
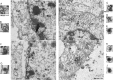
VP8 is a major tegument protein of bovine herpesvirus 1 (BoHV-1) and is essential for viral replication in cattle. The protein undergoes phosphorylation after transcription through cellular casein kinase 2 (CK2) and a viral kinase, US3. In this study, a virus containing a mutated VP8 protein that is not phosphorylated by CK2 and US3 (BoHV-1-YmVP8) was constructed by homologous recombination in mammalian cells. When BoHV-1-YmVP8-infected cells were observed by transmission electron microscopy, blocking phosphorylation of VP8 was found to impair viral DNA encapsidation, resulting in release of incomplete viral particles to the extracellular environment. Consequently, less infectious virus was produced by the mutant virus than by wild-type (WT) virus. A comparison of mutant and WT VP8 by confocal microscopy revealed that mutant VP8 is nuclear throughout infection while WT VP8 is nuclear early during infection and is associated with the Golgi apparatus at later stages. This, together with the observation that mutant VP8 is present in virions, albeit in smaller amounts, suggests that the incorporation of VP8 may occur at two stages. The first takes place without the need for phosphorylation and before or during nuclear egress of capsids, whereas the second occurs in the Golgi apparatus and requires phosphorylation of VP8. The results indicate that phosphorylated VP8 plays a role in viral DNA encapsidation and in the secondary virion incorporation of VP8. To perform these functions, the cellular localization of VP8 is adjusted based on the phosphorylation status.
Importance: In this study, phosphorylation of VP8 was shown to have a function in BoHV-1 replication. A virus containing a mutated VP8 protein that is not phosphorylated by CK2 and US3 (BoHV-1-YmVP8) produced smaller numbers of infectious virions than wild-type (WT) virus. The maturation and egress of WT and mutant BoHV-1 were studied, showing a process similar to that reported for other alphaherpesviruses. Interestingly, lack of phosphorylation of VP8 by CK2 and US3 resulted in reduced incorporation of viral DNA into capsids during mutant BoHV-1 infection, as well as lower numbers of extracellular virions. Furthermore, mutant VP8 remained nuclear throughout infection, in contrast to WT VP8, which is nuclear at early stages and Golgi apparatus associated late during infection. This correlates with smaller amounts of mutant VP8 in virions and suggests for the first time that VP8 may be assembled into the virions at two stages, with the latter dependent on phosphorylation.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
US3 Kinase-Mediated Phosphorylation of Tegument Protein VP8 Plays a Critical Role in the Cellular Localization of VP8 and Its Effect on the Lipid Metabolism of Bovine Herpesvirus 1-Infected Cells.J Virol. 2019 Mar 5;93(6):e02151-18. doi: 10.1128/JVI.02151-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626671 Free PMC article.
-
Regulation and function of phosphorylation on VP8, the major tegument protein of bovine herpesvirus 1.J Virol. 2015 Apr;89(8):4598-611. doi: 10.1128/JVI.03180-14. Epub 2015 Feb 11. J Virol. 2015. PMID: 25673708 Free PMC article.
-
VP8, the Major Tegument Protein of Bovine Herpesvirus 1, Interacts with Cellular STAT1 and Inhibits Interferon Beta Signaling.J Virol. 2016 Apr 29;90(10):4889-4904. doi: 10.1128/JVI.00017-16. Print 2016 May 15. J Virol. 2016. PMID: 26889034 Free PMC article.
-
Bovine Herpesvirus 1 Counteracts Immune Responses and Immune-Surveillance to Enhance Pathogenesis and Virus Transmission.Front Immunol. 2019 May 7;10:1008. doi: 10.3389/fimmu.2019.01008. eCollection 2019. Front Immunol. 2019. PMID: 31134079 Free PMC article. Review.
-
Nuclear Egress of Herpesviruses: The Prototypic Vesicular Nucleocytoplasmic Transport.Adv Virus Res. 2016;94:81-140. doi: 10.1016/bs.aivir.2015.10.002. Epub 2016 Jan 29. Adv Virus Res. 2016. PMID: 26997591 Review.
Cited by
-
Common and Strain-Specific Post-Translational Modifications of the Potyvirus Plum pox virus Coat Protein in Different Hosts.Viruses. 2020 Mar 12;12(3):308. doi: 10.3390/v12030308. Viruses. 2020. PMID: 32178365 Free PMC article.
-
Bovine herpesvirus 1 tegument protein UL21 plays critical roles in viral secondary envelopment and cell-to-cell spreading.Oncotarget. 2017 Oct 10;8(55):94462-94480. doi: 10.18632/oncotarget.21776. eCollection 2017 Nov 7. Oncotarget. 2017. PMID: 29212242 Free PMC article.
-
Structural and Functional Diversity among Five RING Finger Proteins from Carassius Auratus Herpesvirus (CaHV).Viruses. 2021 Feb 7;13(2):254. doi: 10.3390/v13020254. Viruses. 2021. PMID: 33562288 Free PMC article.
-
Duck plague virus US3 protein kinase phosphorylates UL47 and regulates the subcellular localization of UL47.Front Microbiol. 2022 Oct 25;13:876820. doi: 10.3389/fmicb.2022.876820. eCollection 2022. Front Microbiol. 2022. PMID: 36386680 Free PMC article.
-
Bovine Herpesvirus-1 Glycoprotein M Mediates the Translocation to the Golgi Apparatus and Packaging of VP8.Viruses. 2022 Sep 8;14(9):1985. doi: 10.3390/v14091985. Viruses. 2022. PMID: 36146791 Free PMC article.
References
-
- Kato A, Liu Z, Minowa A, Imai T, Tanaka M, Sugimoto K, Nishiyama Y, Arii J, Kawaguchi Y. 2011. Herpes simplex virus 1 protein kinase Us3 and major tegument protein UL47 reciprocally regulate their subcellular localization in infected cells. J Virol 85:9599–9613. doi:10.1128/JVI.00845-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

