Expression of Oncogenic Alleles Induces Multiple Blocks to Human Cytomegalovirus Infection
- PMID: 26889030
- PMCID: PMC4836323
- DOI: 10.1128/JVI.00179-16
Expression of Oncogenic Alleles Induces Multiple Blocks to Human Cytomegalovirus Infection
Abstract
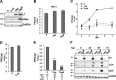
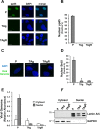
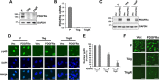
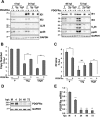
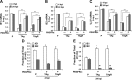
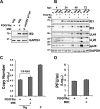
In contrast to many viruses, human cytomegalovirus (HCMV) is unable to productively infect most cancer-derived cell lines. The mechanisms of this restriction are unclear. To explore this issue, we tested whether defined oncogenic alleles, including the simian virus 40 (SV40) T antigen (TAg) and oncogenic H-Ras, inhibit HCMV infection. We found that expression of SV40 TAg blocks HCMV infection in human fibroblasts, whereas the replication of a related herpesvirus, herpes simplex virus 1 (HSV-1), was not impacted. The earliest restriction of HCMV infection involves a block of viral entry, as TAg expression prevented the nuclear delivery of viral DNA and pp65. Subsequently, we found that TAg expression reduces the abundance of platelet-derived growth factor receptor α (PDGFRα), a host protein important for HCMV entry. Viral entry into TAg-immortalized fibroblasts could largely be rescued by PDGFRα overexpression. Similarly, PDGFRα overexpression in HeLa cells markedly increased the levels of HCMV gene expression and DNA replication. However, the robust production of viral progeny was not restored by PDGFRα overexpression in either HeLa cells or TAg-immortalized fibroblasts, suggesting additional restrictions associated with transformation and TAg expression. In TAg-expressing fibroblasts, expression of the immediate early 2 (IE2) protein was not rescued to the same extent as that of the immediate early 1 (IE1) protein, suggesting that TAg expression impacts the accumulation of major immediate early (MIE) transcripts. Transduction of IE2 largely rescued HCMV gene expression in TAg-expressing fibroblasts but did not rescue the production of infectious virions. Collectively, our data indicate that oncogenic alleles induce multiple restrictions to HCMV replication.
Importance: HCMV cannot replicate in most cancerous cells, yet the causes of this restriction are not clear. The mechanisms that restrict viral replication in cancerous cells represent viral vulnerabilities that can potentially be exploited therapeutically in other contexts. Here we found that SV40 T antigen-mediated transformation inhibits HCMV infection at multiple points in the viral life cycle, including through inhibition of proper viral entry, normal expression of immediate early genes, and viral DNA replication. Our results suggest that the SV40 T antigen could be a valuable tool to dissect cellular activities that are important for successful infection, thereby potentially informing novel antiviral development strategies. This is an important consideration, given that HCMV is a leading cause of birth defects and causes severe infection in immunocompromised individuals.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
The 5' Untranslated Region of the Major Immediate Early mRNA Is Necessary for Efficient Human Cytomegalovirus Replication.J Virol. 2018 Mar 14;92(7):e02128-17. doi: 10.1128/JVI.02128-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29343581 Free PMC article.
-
Analysis of the functional interchange between the IE1 and pp71 proteins of human cytomegalovirus and ICP0 of herpes simplex virus 1.J Virol. 2015 Mar;89(6):3062-75. doi: 10.1128/JVI.03480-14. Epub 2014 Dec 31. J Virol. 2015. PMID: 25552717 Free PMC article.
-
Chromatin-Remodeling Factor SPOC1 Acts as a Cellular Restriction Factor against Human Cytomegalovirus by Repressing the Major Immediate Early Promoter.J Virol. 2018 Jun 29;92(14):e00342-18. doi: 10.1128/JVI.00342-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29743358 Free PMC article.
-
Intrinsic Immune Mechanisms Restricting Human Cytomegalovirus Replication.Viruses. 2021 Jan 26;13(2):179. doi: 10.3390/v13020179. Viruses. 2021. PMID: 33530304 Free PMC article. Review.
-
Bright and Early: Inhibiting Human Cytomegalovirus by Targeting Major Immediate-Early Gene Expression or Protein Function.Viruses. 2020 Jan 16;12(1):110. doi: 10.3390/v12010110. Viruses. 2020. PMID: 31963209 Free PMC article. Review.
Cited by
-
Cellular Transformation by Human Cytomegalovirus.Cancers (Basel). 2024 May 22;16(11):1970. doi: 10.3390/cancers16111970. Cancers (Basel). 2024. PMID: 38893091 Free PMC article.
-
Human cytomegalovirus UL23 exploits PD-L1 inhibitory signaling pathway to evade T cell-mediated cytotoxicity.mBio. 2024 Jul 17;15(7):e0119124. doi: 10.1128/mbio.01191-24. Epub 2024 Jun 3. mBio. 2024. PMID: 38829126 Free PMC article.
-
EZH2-Myc driven glioblastoma elicited by cytomegalovirus infection of human astrocytes.Oncogene. 2023 Jun;42(24):2031-2045. doi: 10.1038/s41388-023-02709-3. Epub 2023 May 5. Oncogene. 2023. PMID: 37147437 Free PMC article.
-
Human cytomegalovirus attenuates AKT activity by destabilizing insulin receptor substrate proteins.bioRxiv [Preprint]. 2023 Apr 17:2023.04.17.537203. doi: 10.1101/2023.04.17.537203. bioRxiv. 2023. Update in: J Virol. 2023 Oct 31;97(10):e0056323. doi: 10.1128/jvi.00563-23 PMID: 37131605 Free PMC article. Updated. Preprint.
-
YTHDF2 Is Downregulated in Response to Host Shutoff Induced by DNA Virus Infection and Regulates Interferon-Stimulated Gene Expression.J Virol. 2023 Mar 30;97(3):e0175822. doi: 10.1128/jvi.01758-22. Epub 2023 Mar 14. J Virol. 2023. PMID: 36916936 Free PMC article.
References
-
- Mocarski ES, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2701–2757. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE PM (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

