Novel fusion proteins for the antigen-specific staining and elimination of B cell receptor-positive cell populations demonstrated by a tetanus toxoid fragment C (TTC) model antigen
- PMID: 26883813
- PMCID: PMC4756516
- DOI: 10.1186/s12896-016-0249-x
Novel fusion proteins for the antigen-specific staining and elimination of B cell receptor-positive cell populations demonstrated by a tetanus toxoid fragment C (TTC) model antigen
Abstract
Background: In an earlier study we developed a unique strategy allowing us to specifically eliminate antigen-specific murine B cells via their distinct B cell receptors using a new class of fusion proteins. In the present work we elaborated our idea to demonstrate the feasibility of specifically addressing and eliminating human memory B cells.
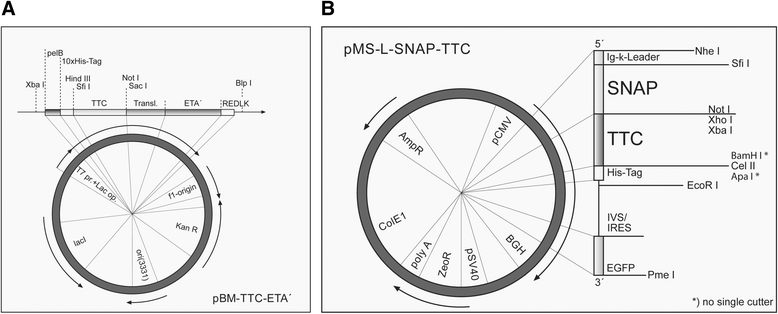
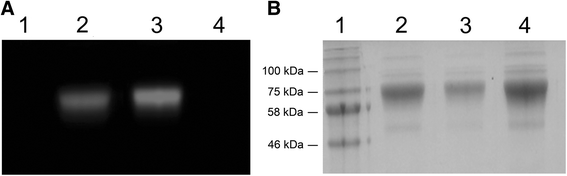
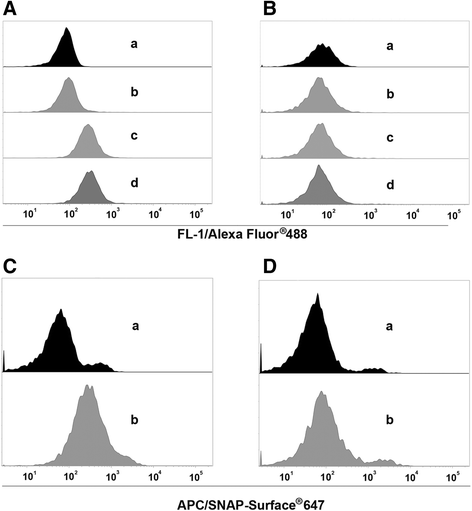
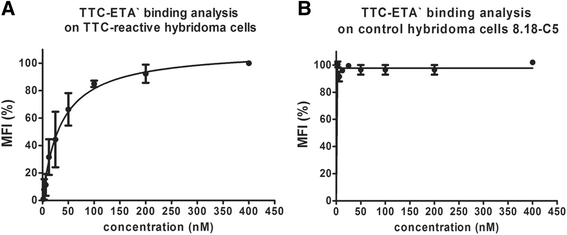
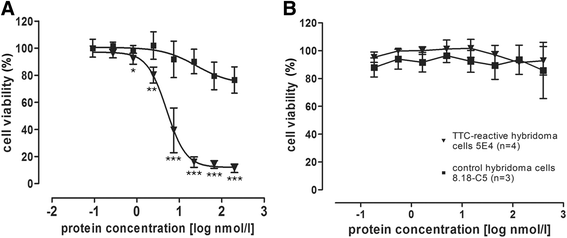
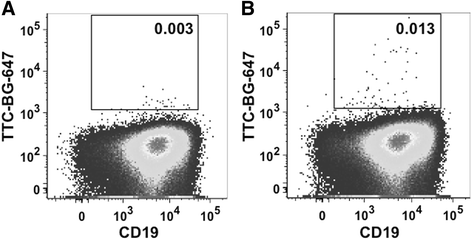
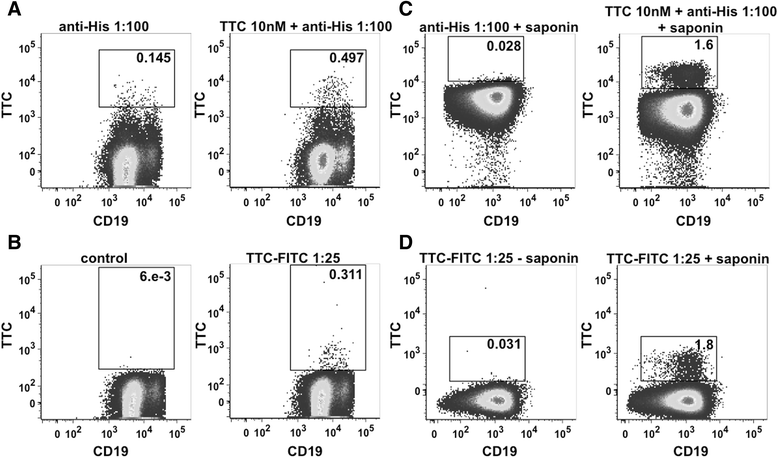
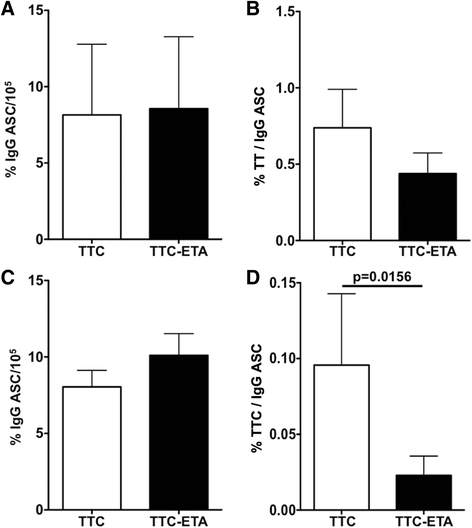
Results: The present study reveals efficient adaptation of the general approach to selectively target and eradicate human memory B cells. In order to demonstrate the feasibility we engineered a fusion protein following the principle of recombinant immunotoxins by combining a model antigen (tetanus toxoid fragment C, TTC) for B cell receptor targeting and a truncated version of Pseudomonas aeruginosa exotoxin A (ETA') to induce apoptosis after cellular uptake. The TTC-ETA' fusion protein not only selectively bound to a TTC-reactive murine B cell hybridoma cell line in vitro but also to freshly isolated human memory B cells from immunized donors ex vivo. Specific toxicity was confirmed on an antigen-specific population of human CD27(+) memory B cells.
Conclusions: This protein engineering strategy can be used as a generalized platform approach for the construction of therapeutic fusion proteins with disease-relevant antigens as B cell receptor-binding domains, offering a promising approach for the specific depletion of autoreactive B-lymphocytes in B cell-driven autoimmune diseases.
Figures
Similar articles
-
Generation of an artificial human B cell line test system using Transpo-mAbTM technology to evaluate the therapeutic efficacy of novel antigen-specific fusion proteins.PLoS One. 2017 Jul 13;12(7):e0180305. doi: 10.1371/journal.pone.0180305. eCollection 2017. PLoS One. 2017. PMID: 28704435 Free PMC article.
-
Depletion of autoreactive B-lymphocytes by a recombinant myelin oligodendrocyte glycoprotein-based immunotoxin.J Neuroimmunol. 2008 Mar;195(1-2):28-35. doi: 10.1016/j.jneuroim.2008.01.001. Epub 2008 Feb 15. J Neuroimmunol. 2008. PMID: 18280586
-
[Gene cloning, optimized expression and immunogenicity evaluation of tetanus toxin fragment C].Nan Fang Yi Ke Da Xue Xue Bao. 2008 May;28(5):731-5. Nan Fang Yi Ke Da Xue Xue Bao. 2008. PMID: 18504192 Chinese.
-
Antigen-specific targeting and elimination of EBV-transformed B cells by allergen toxins.J Allergy Clin Immunol. 2005 Oct;116(4):910-5. doi: 10.1016/j.jaci.2005.07.022. J Allergy Clin Immunol. 2005. PMID: 16210069
-
Selective killing of B-cell hybridomas targeting proteinase 3, Wegener's autoantigen.Immunology. 2004 Jun;112(2):228-36. doi: 10.1111/j.1365-2567.2004.01875.x. Immunology. 2004. PMID: 15147566 Free PMC article.
Cited by
-
Human Antibody Fusion Proteins/Antibody Drug Conjugates in Breast and Ovarian Cancer.Transfus Med Hemother. 2017 Sep;44(5):303-310. doi: 10.1159/000479979. Epub 2017 Sep 11. Transfus Med Hemother. 2017. PMID: 29070975 Free PMC article. Review.
-
Generation of an artificial human B cell line test system using Transpo-mAbTM technology to evaluate the therapeutic efficacy of novel antigen-specific fusion proteins.PLoS One. 2017 Jul 13;12(7):e0180305. doi: 10.1371/journal.pone.0180305. eCollection 2017. PLoS One. 2017. PMID: 28704435 Free PMC article.
-
A recombinant Der p 1-specific allergen-toxin demonstrates superior killing of allergen-reactive IgG+ hybridomas in comparison to its recombinant allergen-drug conjugate.Immunother Adv. 2022 Dec 6;3(1):ltac023. doi: 10.1093/immadv/ltac023. eCollection 2023. Immunother Adv. 2022. PMID: 36789295 Free PMC article.
-
Sequential Prodrug Strategy To Target and Eliminate ACPA-Selective Autoreactive B Cells.Mol Pharm. 2018 Dec 3;15(12):5565-5573. doi: 10.1021/acs.molpharmaceut.8b00741. Epub 2018 Oct 26. Mol Pharm. 2018. PMID: 30289723 Free PMC article.
References
-
- Mamula MJ. B lymphocyte biology in SLE. In: Systemic Lupus Erythematosus. 5th ed., Edited by R.G. Lahita TK, J. Buyon, G. Tsokos. Boston: Elsevier/Academic Press, 2010.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

