Critical POU domain residues confer Oct4 uniqueness in somatic cell reprogramming
- PMID: 26877091
- PMCID: PMC4753506
- DOI: 10.1038/srep20818
Critical POU domain residues confer Oct4 uniqueness in somatic cell reprogramming
Abstract
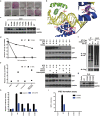
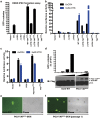
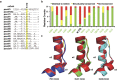
The POU domain transcription factor Oct4 plays critical roles in self-renewal and pluripotency of embryonic stem cells (ESCs). Together with Sox2, Klf4 and c-Myc, Oct4 can reprogram any other cell types to pluripotency, in which Oct4 is the only factor that cannot be functionally replaced by other POU family members. To investigate the determinant elements of Oct4 uniqueness, we performed Ala scan on all Ser, Thr, Tyr, Lys and Arg of murine Oct4 by testing their capability in somatic cell reprogramming. We uncovered a series of residues that are important for Oct4 functionality, in which almost all of these key residues are within the POU domains making direct interaction with DNA. The Oct4 N- and C-terminal transactivation domains (TADs) are not unique and could be replaced by the Yes-associated protein (YAP) TAD domain to support reprogramming. More importantly, we uncovered two important residues that confer Oct4 uniqueness in somatic cell reprogramming. Our systematic structure-function analyses bring novel mechanistic insight into the molecular basis of how critical residues function together to confer Oct4 uniqueness among POU family for somatic cell reprogramming.
Figures


Similar articles
-
Enhanced human somatic cell reprogramming efficiency by fusion of the MYC transactivation domain and OCT4.Stem Cell Res. 2017 Dec;25:88-97. doi: 10.1016/j.scr.2017.10.014. Epub 2017 Oct 26. Stem Cell Res. 2017. PMID: 29125994
-
Optimal reprogramming factor stoichiometry increases colony numbers and affects molecular characteristics of murine induced pluripotent stem cells.Cytometry A. 2011 Jun;79(6):426-35. doi: 10.1002/cyto.a.21072. Epub 2011 May 4. Cytometry A. 2011. PMID: 21548079
-
Manipulation of KLF4 expression generates iPSCs paused at successive stages of reprogramming.Stem Cell Reports. 2014 Nov 11;3(5):915-29. doi: 10.1016/j.stemcr.2014.08.014. Epub 2014 Oct 2. Stem Cell Reports. 2014. PMID: 25418733 Free PMC article.
-
Mechanism of Induction: Induced Pluripotent Stem Cells (iPSCs).J Stem Cells. 2015;10(1):43-62. J Stem Cells. 2015. PMID: 26665937 Review.
-
The genetics of induced pluripotency.Reproduction. 2010 Jan;139(1):35-44. doi: 10.1530/REP-09-0024. Reproduction. 2010. PMID: 19605512 Review.
Cited by
-
Phosphorylation of OCT4 Serine 236 Inhibits Germ Cell Tumor Growth by Inducing Differentiation.Cancers (Basel). 2020 Sep 11;12(9):2601. doi: 10.3390/cancers12092601. Cancers (Basel). 2020. PMID: 32932964 Free PMC article.
-
Permissive epigenomes endow reprogramming competence to transcriptional regulators.Nat Chem Biol. 2021 Jan;17(1):47-56. doi: 10.1038/s41589-020-0618-6. Epub 2020 Aug 17. Nat Chem Biol. 2021. PMID: 32807969
-
POU3F3-related disorder: Defining the phenotype and expanding the molecular spectrum.Clin Genet. 2023 Aug;104(2):186-197. doi: 10.1111/cge.14353. Epub 2023 May 10. Clin Genet. 2023. PMID: 37165752 Free PMC article.
-
Reprogramming competence of OCT factors is determined by transactivation domains.Sci Adv. 2020 Sep 2;6(36):eaaz7364. doi: 10.1126/sciadv.aaz7364. Print 2020 Sep. Sci Adv. 2020. PMID: 32917606 Free PMC article.
-
Oct4 redox sensitivity potentiates reprogramming and differentiation.Genes Dev. 2024 May 21;38(7-8):308-321. doi: 10.1101/gad.351411.123. Genes Dev. 2024. PMID: 38719541 Free PMC article.
References
-
- Downs K. M. Systematic localization of Oct-3/4 to the gastrulating mouse conceptus suggests manifold roles in mammalian development. Dev. Dyn. 237, 464–475 (2008). - PubMed
-
- Takahashi K. & Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). - PubMed
-
- Esch D. et al. A unique Oct4 interface is crucial for reprogramming to pluripotency. Nat. Cell Biol. 15, 295–301 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

