Identifying the effect of patient sharing on between-hospital genetic differentiation of methicillin-resistant Staphylococcus aureus
- PMID: 26873713
- PMCID: PMC4752745
- DOI: 10.1186/s13073-016-0274-3
Identifying the effect of patient sharing on between-hospital genetic differentiation of methicillin-resistant Staphylococcus aureus
Abstract
Background: Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most common healthcare-associated pathogens. To examine the role of inter-hospital patient sharing on MRSA transmission, a previous study collected 2,214 samples from 30 hospitals in Orange County, California and showed by spa typing that genetic differentiation decreased significantly with increased patient sharing. In the current study, we focused on the 986 samples with spa type t008 from the same population.
Methods: We used genome sequencing to determine the effect of patient sharing on genetic differentiation between hospitals. Genetic differentiation was measured by between-hospital genetic diversity, F ST , and the proportion of nearly identical isolates between hospitals.
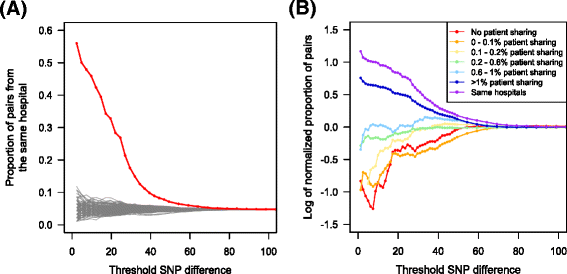
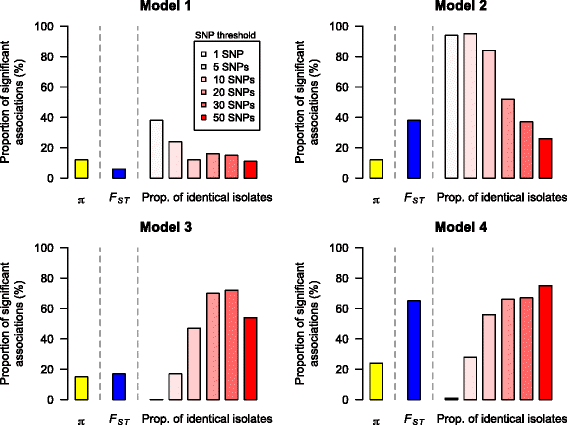
Results: Surprisingly, we found very similar genetic diversity within and between hospitals, and no significant association between patient sharing and genetic differentiation measured by F ST . However, in contrast to F ST , there was a significant association between patient sharing and the proportion of nearly identical isolates between hospitals. We propose that the proportion of nearly identical isolates is more powerful at determining transmission dynamics than traditional estimators of genetic differentiation (F ST ) when gene flow between populations is high, since it is more responsive to recent transmission events. Our hypothesis was supported by the results from coalescent simulations.
Conclusions: Our results suggested that there was a high level of gene flow between hospitals facilitated by patient sharing, and that the proportion of nearly identical isolates is more sensitive to population structure than F ST when gene flow is high.
Figures
Similar articles
-
Frequent Undetected Ward-Based Methicillin-Resistant Staphylococcus aureus Transmission Linked to Patient Sharing Between Hospitals.Clin Infect Dis. 2018 Mar 5;66(6):840-848. doi: 10.1093/cid/cix901. Clin Infect Dis. 2018. PMID: 29095965 Free PMC article.
-
Molecular characterization of Staphylococcus aureus isolates from various healthcare institutions in Nairobi, Kenya: a cross sectional study.Ann Clin Microbiol Antimicrob. 2016 Sep 20;15(1):51. doi: 10.1186/s12941-016-0171-z. Ann Clin Microbiol Antimicrob. 2016. PMID: 27647271 Free PMC article.
-
Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus (ST398) in Dutch hospitals.Clin Microbiol Infect. 2011 Feb;17(2):316-9. doi: 10.1111/j.1469-0691.2010.03260.x. Clin Microbiol Infect. 2011. PMID: 20459436
-
Molecular analysis of methicillin-resistant Staphylococcus aureus dissemination among healthcare professionals and/or HIV patients from a tertiary hospital.Rev Soc Bras Med Trop. 2016 Feb;49(1):51-6. doi: 10.1590/0037-8682-0284-2015. Rev Soc Bras Med Trop. 2016. PMID: 27163564
-
[Infectivity-resistotype-genotype clustering of methicillin-resistant Staphylococcus aureus strains in the Central Blacksea Region of Turkey].Mikrobiyol Bul. 2014 Jan;48(1):14-27. Mikrobiyol Bul. 2014. PMID: 24506712 Turkish.
Cited by
-
Mapping imported malaria in Bangladesh using parasite genetic and human mobility data.Elife. 2019 Apr 2;8:e43481. doi: 10.7554/eLife.43481. Elife. 2019. PMID: 30938289 Free PMC article.
-
Widespread sharing of pneumococcal strains in a rural African setting: proximate villages are more likely to share similar strains that are carried at multiple timepoints.Microb Genom. 2022 Feb;8(2):000732. doi: 10.1099/mgen.0.000732. Microb Genom. 2022. PMID: 35119356 Free PMC article.
-
Range Expansion and the Origin of USA300 North American Epidemic Methicillin-Resistant Staphylococcus aureus.mBio. 2018 Jan 2;9(1):e02016-17. doi: 10.1128/mBio.02016-17. mBio. 2018. PMID: 29295910 Free PMC article.
-
Threshold-free genomic cluster detection to track transmission pathways in health-care settings: a genomic epidemiology analysis.Lancet Microbe. 2022 Sep;3(9):e652-e662. doi: 10.1016/S2666-5247(22)00115-X. Epub 2022 Jul 5. Lancet Microbe. 2022. PMID: 35803292 Free PMC article.
-
Mapping malaria by combining parasite genomic and epidemiologic data.BMC Med. 2018 Oct 18;16(1):190. doi: 10.1186/s12916-018-1181-9. BMC Med. 2018. PMID: 30333020 Free PMC article.
References
-
- Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol. 2013;34:1–14. doi: 10.1086/668770. - DOI - PubMed
-
- Centers for Disease Control and Prevention . Active Bacterial Core Surveillance Report, Emerging Infections Program Network, Methicillin-Resistant Staphylococcus aureus, 2012. Atlanta, GA: CDC; 2012.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

