Drosophila Females Undergo Genome Expansion after Interspecific Hybridization
- PMID: 26872773
- PMCID: PMC4824032
- DOI: 10.1093/gbe/evw024
Drosophila Females Undergo Genome Expansion after Interspecific Hybridization
Abstract
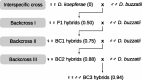
Genome size (or C-value) can present a wide range of values among eukaryotes. This variation has been attributed to differences in the amplification and deletion of different noncoding repetitive sequences, particularly transposable elements (TEs). TEs can be activated under different stress conditions such as interspecific hybridization events, as described for several species of animals and plants. These massive transposition episodes can lead to considerable genome expansions that could ultimately be involved in hybrid speciation processes. Here, we describe the effects of hybridization and introgression on genome size of Drosophila hybrids. We measured the genome size of two close Drosophila species, Drosophila buzzatii and Drosophila koepferae, their F1 offspring and the offspring from three generations of backcrossed hybrids; where mobilization of up to 28 different TEs was previously detected. We show that hybrid females indeed present a genome expansion, especially in the first backcross, which could likely be explained by transposition events. Hybrid males, which exhibit more variable C-values among individuals of the same generation, do not present an increased genome size. Thus, we demonstrate that the impact of hybridization on genome size can be detected through flow cytometry and is sex-dependent.
Keywords: AFLP markers; Drosophila; flow cytometry; genome size; hybrids; transposable elements.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
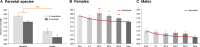
Similar articles
-
Changes of Osvaldo expression patterns in germline of male hybrids between the species Drosophila buzzatii and Drosophila koepferae.Mol Genet Genomics. 2015 Aug;290(4):1471-83. doi: 10.1007/s00438-015-1012-z. Epub 2015 Feb 25. Mol Genet Genomics. 2015. PMID: 25711309
-
Transposable Element Misregulation Is Linked to the Divergence between Parental piRNA Pathways in Drosophila Hybrids.Genome Biol Evol. 2017 Jun 1;9(6):1450-1470. doi: 10.1093/gbe/evx091. Genome Biol Evol. 2017. PMID: 28854624 Free PMC article.
-
Adaptation of the AFLP technique as a new tool to detect genetic instability and transposition in interspecific hybrids.Biotechniques. 2011 Apr;50(4):247-50. doi: 10.2144/000113655. Biotechniques. 2011. PMID: 21548908
-
What transposable elements tell us about genome organization and evolution: the case of Drosophila.Cytogenet Genome Res. 2005;110(1-4):25-34. doi: 10.1159/000084935. Cytogenet Genome Res. 2005. PMID: 16093655 Review.
-
The challenges of predicting transposable element activity in hybrids.Curr Genet. 2021 Aug;67(4):567-572. doi: 10.1007/s00294-021-01169-0. Epub 2021 Mar 18. Curr Genet. 2021. PMID: 33738571 Review.
Cited by
-
Drosophila Interspecific Hybridization Causes A Deregulation of the piRNA Pathway Genes.Genes (Basel). 2020 Feb 19;11(2):215. doi: 10.3390/genes11020215. Genes (Basel). 2020. PMID: 32092860 Free PMC article.
-
A Transposon Story: From TE Content to TE Dynamic Invasion of Drosophila Genomes Using the Single-Molecule Sequencing Technology from Oxford Nanopore.Cells. 2020 Jul 25;9(8):1776. doi: 10.3390/cells9081776. Cells. 2020. PMID: 32722451 Free PMC article.
-
Dissecting the Satellite DNA Landscape in Three Cactophilic Drosophila Sequenced Genomes.G3 (Bethesda). 2017 Aug 7;7(8):2831-2843. doi: 10.1534/g3.117.042093. G3 (Bethesda). 2017. PMID: 28659292 Free PMC article.
-
High Stability of the Epigenome in Drosophila Interspecific Hybrids.Genome Biol Evol. 2022 Feb 4;14(2):evac024. doi: 10.1093/gbe/evac024. Genome Biol Evol. 2022. PMID: 35143649 Free PMC article.
-
Living Organisms Author Their Read-Write Genomes in Evolution.Biology (Basel). 2017 Dec 6;6(4):42. doi: 10.3390/biology6040042. Biology (Basel). 2017. PMID: 29211049 Free PMC article. Review.
References
-
- Boulesteix M, Weiss M, Biémont C. 2006. Differences in genome size between closely related species: the Drosophila melanogaster species subgroup. Mol Biol Evol. 23:162–167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

