Reduced Ets Domain-containing Protein Elk1 Promotes Pulmonary Fibrosis via Increased Integrin αvβ6 Expression
- PMID: 26861876
- PMCID: PMC4850293
- DOI: 10.1074/jbc.M115.692368
Reduced Ets Domain-containing Protein Elk1 Promotes Pulmonary Fibrosis via Increased Integrin αvβ6 Expression
Abstract
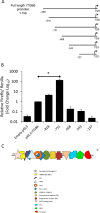
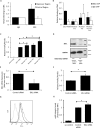
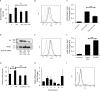
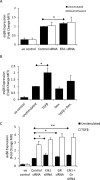
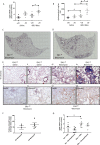
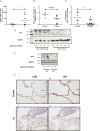
Idiopathic pulmonary fibrosis (IPF) is a progressive fibrotic lung disease with high mortality. Active TGFβ1 is considered central to the pathogenesis of IPF. A major mechanism of TGFβ1 activation in the lung involves the epithelially restricted αvβ6 integrin. Expression of the αvβ6 integrin is dramatically increased in IPF. How αvβ6 integrin expression is regulated in the pulmonary epithelium is unknown. Here we identify a region in the β6 subunit gene (ITGB6) promoter acting to markedly repress basal gene transcription, which responds to both the Ets domain-containing protein Elk1 (Elk1) and the glucocorticoid receptor (GR). Both Elk1 and GR can regulate αvβ6 integrin expression in vitro We demonstrate Elk1 binding to the ITGB6 promoter basally and that manipulation of Elk1 or Elk1 binding alters ITGB6 promoter activity, gene transcription, and αvβ6 integrin expression. Crucially, we find that loss of Elk1 causes enhanced Itgb6 expression and exaggerated lung fibrosis in an in vivo model of fibrosis, whereas the GR agonist dexamethasone inhibits Itgb6 expression. Moreover, Elk1 dysregulation is present in epithelium from patients with IPF. These data reveal a novel role for Elk1 regulating ITGB6 expression and highlight how dysregulation of Elk1 can contribute to human disease.
Keywords: elk1; fibrosis; gene regulation; integrin; lung; pulmonary fibrosis.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
Similar articles
-
Amplification of TGFβ Induced ITGB6 Gene Transcription May Promote Pulmonary Fibrosis.PLoS One. 2016 Aug 5;11(8):e0158047. doi: 10.1371/journal.pone.0158047. eCollection 2016. PLoS One. 2016. PMID: 27494713 Free PMC article.
-
Loss of ELK1 has differential effects on age-dependent organ fibrosis.Int J Biochem Cell Biol. 2020 Mar;120:105668. doi: 10.1016/j.biocel.2019.105668. Epub 2019 Dec 23. Int J Biochem Cell Biol. 2020. PMID: 31877385 Free PMC article.
-
Inhibition of integrin alpha(v)beta6, an activator of latent transforming growth factor-beta, prevents radiation-induced lung fibrosis.Am J Respir Crit Care Med. 2008 Jan 1;177(1):82-90. doi: 10.1164/rccm.200706-806OC. Epub 2007 Oct 4. Am J Respir Crit Care Med. 2008. PMID: 17916808 Free PMC article.
-
The ITGB6 gene: its role in experimental and clinical biology.Gene. 2020 Dec;763S:100023. doi: 10.1016/j.gene.2019.100023. Epub 2019 Nov 6. Gene. 2020. PMID: 34493369 Review.
-
Exploring the role of ITGB6: fibrosis, cancer, and other diseases.Apoptosis. 2024 Jun;29(5-6):570-585. doi: 10.1007/s10495-023-01921-6. Epub 2023 Dec 21. Apoptosis. 2024. PMID: 38127283 Review.
Cited by
-
Association of Human iPSC Gene Signatures and X Chromosome Dosage with Two Distinct Cardiac Differentiation Trajectories.Stem Cell Reports. 2019 Nov 12;13(5):924-938. doi: 10.1016/j.stemcr.2019.09.011. Epub 2019 Oct 24. Stem Cell Reports. 2019. PMID: 31668852 Free PMC article.
-
STAT3 Partly Inhibits Cell Proliferation via Direct Negative Regulation of FST Gene Expression.Front Genet. 2021 Jun 22;12:678667. doi: 10.3389/fgene.2021.678667. eCollection 2021. Front Genet. 2021. PMID: 34239543 Free PMC article.
-
Lysyl oxidase like 2 is increased in asthma and contributes to asthmatic airway remodelling.Eur Respir J. 2022 Jul 7;60(1):2004361. doi: 10.1183/13993003.04361-2020. Print 2022 Jul. Eur Respir J. 2022. PMID: 34996828 Free PMC article.
-
Integrin subunit beta 6 is a potential diagnostic marker for acute kidney injury in patients with diabetic kidney disease: a single cell sequencing data analysis.Ren Fail. 2024 Dec;46(2):2409348. doi: 10.1080/0886022X.2024.2409348. Epub 2024 Oct 2. Ren Fail. 2024. PMID: 39356055 Free PMC article.
-
Knockdown of lncRNA LINC00662 suppresses malignant behaviour of osteosarcoma cells via competition with miR-30b-3p to regulate ELK1 expression.J Orthop Surg Res. 2022 Feb 5;17(1):74. doi: 10.1186/s13018-022-02964-2. J Orthop Surg Res. 2022. PMID: 35123530 Free PMC article.
References
-
- Vancheri C., Failla M., Crimi N., and Raghu G. (2010) Idiopathic pulmonary fibrosis: a disease with similarities and links to cancer biology. Eur. Respir. J. 35, 496–504 - PubMed
-
- Navaratnam V., Fleming K. M., West J., Smith C. J., Jenkins R. G., Fogarty A., and Hubbard R. B. (2011) The rising incidence of idiopathic pulmonary fibrosis in the U.K. Thorax 66, 462–467 - PubMed
-
- Tatler A. L., and Jenkins G. (2012) TGF-β activation and lung fibrosis. Proc. Am. Thorac. Soc. 9, 130–136 - PubMed
-
- Hagimoto N., Kuwano K., Inoshima I., Yoshimi M., Nakamura N., Fujita M., Maeyama T., and Hara N. (2002) TGF-β 1 as an enhancer of Fas-mediated apoptosis of lung epithelial cells. J. Immunol. 168, 6470–6478 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MR/N005953/1/MRC_/Medical Research Council/United Kingdom
- 085350/WT_/Wellcome Trust/United Kingdom
- G1100564/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- NC/K500501/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- MRF_MRF-091-0004-RG-TATLE/MRF/MRF/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

