Comparison of design strategies for α-helix backbone modification in a protein tertiary fold
- PMID: 26853882
- PMCID: PMC4767680
- DOI: 10.1039/c6cc00273k
Comparison of design strategies for α-helix backbone modification in a protein tertiary fold
Abstract
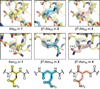
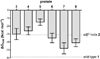
We report here the comparison of five classes of unnatural amino acid building blocks for their ability to be accommodated into an α-helix in a protein tertiary fold context. High-resolution structural characterization and analysis of folding thermodynamics yield new insights into the relationship between backbone composition and folding energetics in α-helix mimetics and suggest refined design rules for engineering the backbones of natural sequences.
Figures


Similar articles
-
Protein-like tertiary folding behavior from heterogeneous backbones.J Am Chem Soc. 2013 Aug 28;135(34):12528-31. doi: 10.1021/ja405422v. Epub 2013 Aug 15. J Am Chem Soc. 2013. PMID: 23937097 Free PMC article.
-
Comparison of backbone modification in protein β-sheets by α→γ residue replacement and α-residue methylation.Org Biomol Chem. 2014 Aug 7;12(29):5375-81. doi: 10.1039/c4ob00886c. Org Biomol Chem. 2014. PMID: 24909436 Free PMC article.
-
Foldamer Tertiary Structure through Sequence-Guided Protein Backbone Alteration.Acc Chem Res. 2018 May 15;51(5):1220-1228. doi: 10.1021/acs.accounts.8b00048. Epub 2018 Apr 19. Acc Chem Res. 2018. PMID: 29672021 Free PMC article.
-
Analysis of folded structure and folding thermodynamics in heterogeneous-backbone proteomimetics.Methods Enzymol. 2021;656:93-122. doi: 10.1016/bs.mie.2021.04.009. Epub 2021 May 3. Methods Enzymol. 2021. PMID: 34325801 Free PMC article. Review.
-
Is protein folding hierarchic? II. Folding intermediates and transition states.Trends Biochem Sci. 1999 Feb;24(2):77-83. doi: 10.1016/s0968-0004(98)01345-0. Trends Biochem Sci. 1999. PMID: 10098403 Review.
Cited by
-
Implications of the unfolded state in the folding energetics of heterogeneous-backbone protein mimetics.Chem Sci. 2022 Sep 20;13(40):11798-11806. doi: 10.1039/d2sc04427g. eCollection 2022 Oct 19. Chem Sci. 2022. PMID: 36320921 Free PMC article.
-
Receptor selectivity from minimal backbone modification of a polypeptide agonist.Proc Natl Acad Sci U S A. 2018 Dec 4;115(49):12383-12388. doi: 10.1073/pnas.1815294115. Epub 2018 Nov 15. Proc Natl Acad Sci U S A. 2018. PMID: 30442659 Free PMC article.
-
Differential Effects of β3 - versus β2 -Amino Acid Residues on the Helicity and Recognition Properties of Bim BH3-Derived α/β-Peptides.Angew Chem Int Ed Engl. 2018 Oct 15;57(42):13829-13832. doi: 10.1002/anie.201806909. Epub 2018 Sep 20. Angew Chem Int Ed Engl. 2018. PMID: 30161284 Free PMC article.
-
Vagabond: bond-based parametrization reduces overfitting for refinement of proteins.Acta Crystallogr D Struct Biol. 2021 Apr 1;77(Pt 4):424-437. doi: 10.1107/S2059798321000826. Epub 2021 Mar 30. Acta Crystallogr D Struct Biol. 2021. PMID: 33825703 Free PMC article.
-
Heterogeneous-Backbone Foldamer Mimics of a Computationally Designed, Disulfide-Rich Miniprotein.Chembiochem. 2019 Jan 2;20(1):103-110. doi: 10.1002/cbic.201800558. Epub 2018 Nov 27. Chembiochem. 2019. PMID: 30326175 Free PMC article.
References
-
- Gellman SH. Acc. Chem. Res. 1998;31:173–180.
-
- Bautista AD, Craig CJ, Harker EA, Schepartz A. Curr. Opin. Chem. Biol. 2007;11:685–692. - PMC - PubMed
- Goodman CM, Choi S, Shandler S, DeGrado WF. Nat. Chem. Biol. 2007;3:252–262. - PMC - PubMed
- Guichard G, Huc I. Chem. Commun. 2011;47:5933–5941. - PubMed
- Horne WS. Expert Opin. Drug Discov. 2011;6:1247–1262. - PubMed
- Martinek TA, Fulop F. Chem. Soc. Rev. 2012;41:687–702. - PubMed
- Sun J, Zuckermann RN. ACS Nano. 2013;7:4715–4732. - PubMed
-
- Hegedüs Z, Wéber E, Kriston-Pál É, Makra I, Czibula Á, Monostori É, Martinek TA. J. Am. Chem. Soc. 2013;135:16578–16584. - PubMed
- Wang PSP, Nguyen JB, Schepartz A. J. Am. Chem. Soc. 2014;136:6810–6813. - PubMed
- Checco JW, Kreitler DF, Thomas NC, Belair DG, Rettko NJ, Murphy WL, Forest KT, Gellman SH. Proc. Natl. Acad. Sci. USA. 2015;112:4552–4557. - PMC - PubMed
-
- Mannige RV, Haxton TK, Proulx C, Robertson EJ, Battigelli A, Butterfoss GL, Zuckermann RN, Whitelam S. Nature. 2015;526:415–420. - PubMed
- Chandramouli N, Ferrand Y, Lautrette G, Kauffmann B, Mackereth CD, Laguerre M, Dubreuil D, Huc I. Nature Chem. 2015;7:334–341. - PubMed
- Collie GW, Pulka-Ziach K, Lombardo CM, Fremaux J, Rosu F, Decossas M, Mauran L, Lambert O, Gabelica V, Mackereth CD, Guichard G. Nature Chem. 2015;7:871–878. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

