Transcription profile of Escherichia coli: genomic SELEX search for regulatory targets of transcription factors
- PMID: 26843427
- PMCID: PMC4797297
- DOI: 10.1093/nar/gkw051
Transcription profile of Escherichia coli: genomic SELEX search for regulatory targets of transcription factors
Abstract
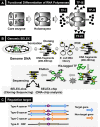
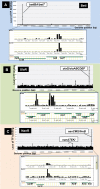
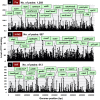
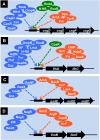
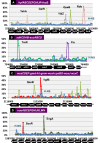
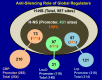
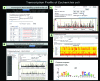
Bacterial genomes are transcribed by DNA-dependent RNA polymerase (RNAP), which achieves gene selectivity through interaction with sigma factors that recognize promoters, and transcription factors (TFs) that control the activity and specificity of RNAP holoenzyme. To understand the molecular mechanisms of transcriptional regulation, the identification of regulatory targets is needed for all these factors. We then performed genomic SELEX screenings of targets under the control of each sigma factor and each TF. Here we describe the assembly of 156 SELEX patterns of a total of 116 TFs performed in the presence and absence of effector ligands. The results reveal several novel concepts: (i) each TF regulates more targets than hitherto recognized; (ii) each promoter is regulated by more TFs than hitherto recognized; and (iii) the binding sites of some TFs are located within operons and even inside open reading frames. The binding sites of a set of global regulators, including cAMP receptor protein, LeuO and Lrp, overlap with those of the silencer H-NS, suggesting that certain global regulators play an anti-silencing role. To facilitate sharing of these accumulated SELEX datasets with the research community, we compiled a database, 'Transcription Profile of Escherichia coli' (www.shigen.nig.ac.jp/ecoli/tec/).
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Integration of regulatory signals through involvement of multiple global regulators: control of the Escherichia coli gltBDF operon by Lrp, IHF, Crp, and ArgR.BMC Microbiol. 2007 Jan 18;7:2. doi: 10.1186/1471-2180-7-2. BMC Microbiol. 2007. PMID: 17233899 Free PMC article.
-
Molecular analysis of the regulation of csiD, a carbon starvation-inducible gene in Escherichia coli that is exclusively dependent on sigma s and requires activation by cAMP-CRP.J Mol Biol. 1998 Feb 20;276(2):339-53. doi: 10.1006/jmbi.1997.1533. J Mol Biol. 1998. PMID: 9512707
-
Genomic SELEX Screening of Regulatory Targets of Escherichia coli Transcription Factors.Methods Mol Biol. 2018;1837:49-69. doi: 10.1007/978-1-4939-8675-0_4. Methods Mol Biol. 2018. PMID: 30109605
-
Building a complete image of genome regulation in the model organism Escherichia coli.J Gen Appl Microbiol. 2018 Jan 15;63(6):311-324. doi: 10.2323/jgam.2017.01.002. Epub 2017 Sep 12. J Gen Appl Microbiol. 2018. PMID: 28904250 Review.
-
Transcription activation by catabolite activator protein (CAP).J Mol Biol. 1999 Oct 22;293(2):199-213. doi: 10.1006/jmbi.1999.3161. J Mol Biol. 1999. PMID: 10550204 Review.
Cited by
-
Fructose-1-kinase has pleiotropic roles in Escherichia coli.bioRxiv [Preprint]. 2023 Dec 14:2023.12.14.571569. doi: 10.1101/2023.12.14.571569. bioRxiv. 2023. Update in: J Biol Chem. 2024 Jun;300(6):107352. doi: 10.1016/j.jbc.2024.107352. PMID: 38168282 Free PMC article. Updated. Preprint.
-
Tunable transcription factor library for robust quantification of regulatory properties in Escherichia coli.Mol Syst Biol. 2022 Jun;18(6):e10843. doi: 10.15252/msb.202110843. Mol Syst Biol. 2022. PMID: 35694815 Free PMC article.
-
Molecular basis for the differential expression of the global regulator VieA in Vibrio cholerae biotypes directed by H-NS, LeuO and quorum sensing.Mol Microbiol. 2018 Feb;107(3):330-343. doi: 10.1111/mmi.13884. Epub 2017 Dec 11. Mol Microbiol. 2018. PMID: 29152799 Free PMC article.
-
The nucleoid protein Dps binds genomic DNA of Escherichia coli in a non-random manner.PLoS One. 2017 Aug 11;12(8):e0182800. doi: 10.1371/journal.pone.0182800. eCollection 2017. PLoS One. 2017. PMID: 28800583 Free PMC article.
-
Exploring the mono-/bistability range of positively autoregulated signaling systems in the presence of competing transcription factor binding sites.PLoS Comput Biol. 2022 Nov 22;18(11):e1010738. doi: 10.1371/journal.pcbi.1010738. eCollection 2022 Nov. PLoS Comput Biol. 2022. PMID: 36413575 Free PMC article.
References
-
- Ishihama A. Prokaryotic genome regulation: multi-factor promoters, multi-target regulators and hierarchic networks. FEMS Microbiol. Rev. 2010;34:628–645. - PubMed
-
- Helmann J., Chamberlin M. Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 1988;57:839–872. - PubMed
-
- Gross C.A., Lonetto M., Losick R. Bacterial sigma factors. In: McKnight SL, Yamamoto KR, editors. Transcriptional Regulation. NY: Cold Spring Harbour Press; 1992. pp. 129–176.
-
- Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 2000;54:499–518. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

