Strand-biased cytosine deamination at the replication fork causes cytosine to thymine mutations in Escherichia coli
- PMID: 26839411
- PMCID: PMC4776466
- DOI: 10.1073/pnas.1522325113
Strand-biased cytosine deamination at the replication fork causes cytosine to thymine mutations in Escherichia coli
Abstract
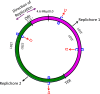
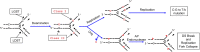
The rate of cytosine deamination is much higher in single-stranded DNA (ssDNA) than in double-stranded DNA, and copying the resulting uracils causes C to T mutations. To study this phenomenon, the catalytic domain of APOBEC3G (A3G-CTD), an ssDNA-specific cytosine deaminase, was expressed in an Escherichia coli strain defective in uracil repair (ung mutant), and the mutations that accumulated over thousands of generations were determined by whole-genome sequencing. C:G to T:A transitions dominated, with significantly more cytosines mutated to thymine in the lagging-strand template (LGST) than in the leading-strand template (LDST). This strand bias was present in both repair-defective and repair-proficient cells and was strongest and highly significant in cells expressing A3G-CTD. These results show that the LGST is accessible to cellular cytosine deaminating agents, explains the well-known GC skew in microbial genomes, and suggests the APOBEC3 family of mutators may target the LGST in the human genome.
Keywords: APOBEC3A; APOBEC3B; cancer genome mutations; kataegis; uracil-DNA glycosylase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

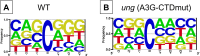
Similar articles
-
Genome-wide mapping of regions preferentially targeted by the human DNA-cytosine deaminase APOBEC3A using uracil-DNA pulldown and sequencing.J Biol Chem. 2019 Oct 11;294(41):15037-15051. doi: 10.1074/jbc.RA119.008053. Epub 2019 Aug 19. J Biol Chem. 2019. PMID: 31431505 Free PMC article.
-
Competition for DNA binding between the genome protector replication protein A and the genome modifying APOBEC3 single-stranded DNA deaminases.Nucleic Acids Res. 2022 Nov 28;50(21):12039-12057. doi: 10.1093/nar/gkac1121. Nucleic Acids Res. 2022. PMID: 36444883 Free PMC article.
-
Relative contribution of cytosine deamination and error-prone replication to the induction of propanodeoxyguanosine-->deoxyadenosine mutations in Escherichia coli.Chem Res Toxicol. 1996 Jan-Feb;9(1):277-83. doi: 10.1021/tx950060w. Chem Res Toxicol. 1996. PMID: 8924603
-
Error-free versus mutagenic processing of genomic uracil--relevance to cancer.DNA Repair (Amst). 2014 Jul;19:38-47. doi: 10.1016/j.dnarep.2014.03.028. Epub 2014 Apr 18. DNA Repair (Amst). 2014. PMID: 24746924 Review.
-
DNA-cytosine deaminases: from antibody maturation to antiviral defense.DNA Repair (Amst). 2004 Jan 5;3(1):85-9. doi: 10.1016/j.dnarep.2003.09.008. DNA Repair (Amst). 2004. PMID: 14697763 Free PMC article. Review.
Cited by
-
A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing.Nature. 2020 Jul;583(7817):631-637. doi: 10.1038/s41586-020-2477-4. Epub 2020 Jul 8. Nature. 2020. PMID: 32641830 Free PMC article.
-
The Effects of a High Concentration of Dissolved Oxygen on Actinobacteria from Lake Baikal.Metabolites. 2023 Jul 7;13(7):830. doi: 10.3390/metabo13070830. Metabolites. 2023. PMID: 37512537 Free PMC article.
-
Evolutionary effects of the AID/APOBEC family of mutagenic enzymes on human gamma-herpesviruses.Virus Evol. 2019 Feb 11;5(1):vey040. doi: 10.1093/ve/vey040. eCollection 2019 Jan. Virus Evol. 2019. PMID: 30792902 Free PMC article.
-
Optimization of amino acid replacement costs by mutational pressure in bacterial genomes.Sci Rep. 2017 Apr 21;7(1):1061. doi: 10.1038/s41598-017-01130-7. Sci Rep. 2017. PMID: 28432324 Free PMC article.
-
Elevated APOBEC3B expression drives a kataegic-like mutation signature and replication stress-related therapeutic vulnerabilities in p53-defective cells.Br J Cancer. 2017 Jun 27;117(1):113-123. doi: 10.1038/bjc.2017.133. Epub 2017 May 23. Br J Cancer. 2017. PMID: 28535155 Free PMC article.
References
-
- Hayatsu H. Bisulfite modification of nucleic acids and their constituents. Prog Nucleic Acid Res Mol Biol. 1976;16:75–124. - PubMed
-
- Paleček E, Bartošík M. Electrochemistry of nucleic acids. Chem Rev. 2012;112(6):3427–3481. - PubMed
-
- Sinden RR. DNA Structure and Function. Academic Press; San Diego, CA: 1994. p. 398.
-
- Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens. Plenum Press; New York, NY: 1983. p. 347.
-
- Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: Determination of rate constants and the activation energy. Biochemistry. 1990;29(10):2532–2537. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

