Physical evidence supporting a ribosomal shunting mechanism of translation initiation for BACE1 mRNA
- PMID: 26824018
- PMCID: PMC4718059
- DOI: 10.4161/trla.24400
Physical evidence supporting a ribosomal shunting mechanism of translation initiation for BACE1 mRNA
Abstract
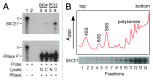
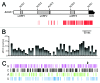
In Alzheimer disease, elevated levels of the BACE1 enzyme are correlated with increased production of amyloid peptides and disease pathology. The increase in BACE1 levels is post-transcriptional and may involve altered translation efficiency. Earlier studies have indicated that translation of BACE1 mRNA is cap-dependent. As ribosomal subunits move from the cap-structure to the initiation codon, they fail to recognize several AUG codons in the 5' leader. In this study, we looked for physical evidence of the mechanism underlying ribosomal scanning or shunting along the BACE1 5' leader by investigating structural stability in the 5' leaders of endogenous mRNAs in vivo. To perform this analysis, we probed RNAs using lead(II) acetate, a cell-permeable chemical that induces cleavage of unpaired nucleotides having conformational flexibility. The data revealed that the ≈440-nt 5' leader was generally resistant to cleavage except for a region upstream of the initiation codon. Cleavage continued into the coding region, consistent with destabilization of secondary structures by translating ribosomes. Evidence that a large segment of the BACE1 5' leader was not cleaved indicates that this region is structurally stable and suggests that it is not scanned. The data support a mechanism of translation initiation in which ribosomal subunits bypass (shunt) part of the BACE1 5' leader to reach the initiation codon. We suggest that a nucleotide bias in the 5' leader may predispose the initiation codon to be more accessible than other AUG codons in the 5' leader, leading to an increase in its relative utilization.
Keywords: BACE1; Initiation; RNase P; Ribosome; Shunt; Translation; mRNA.
Figures


Similar articles
-
Differential utilization of upstream AUGs in the beta-secretase mRNA suggests that a shunting mechanism regulates translation.Proc Natl Acad Sci U S A. 2004 Mar 2;101(9):2794-9. doi: 10.1073/pnas.0308576101. Epub 2004 Feb 23. Proc Natl Acad Sci U S A. 2004. PMID: 14981268 Free PMC article.
-
Reconciling contradictory reports regarding translation of BACE1 mRNA: initiation mechanism is altered by different expression systems.RNA Biol. 2009 Jan-Mar;6(1):54-8. doi: 10.4161/rna.6.1.7567. Epub 2009 Jan 5. RNA Biol. 2009. PMID: 19106624
-
The sequence context of the initiation codon in the encephalomyocarditis virus leader modulates efficiency of internal translation initiation.J Virol. 1992 Apr;66(4):1924-32. doi: 10.1128/JVI.66.4.1924-1932.1992. J Virol. 1992. PMID: 1312611 Free PMC article.
-
Cap-dependent, scanning-free translation initiation mechanisms.Biochim Biophys Acta. 2015 Nov;1849(11):1313-8. doi: 10.1016/j.bbagrm.2015.09.006. Epub 2015 Sep 14. Biochim Biophys Acta. 2015. PMID: 26381322 Review.
-
Structural Insights into the Mechanism of Scanning and Start Codon Recognition in Eukaryotic Translation Initiation.Trends Biochem Sci. 2017 Aug;42(8):589-611. doi: 10.1016/j.tibs.2017.03.004. Epub 2017 Apr 22. Trends Biochem Sci. 2017. PMID: 28442192 Review.
Cited by
-
Cellular mRNA recruits the ribosome via eIF3-PABP bridge to initiate internal translation.RNA Biol. 2017 May 4;14(5):553-567. doi: 10.1080/15476286.2015.1137419. Epub 2016 Feb 1. RNA Biol. 2017. PMID: 26828225 Free PMC article.
-
Role of Eukaryotic Initiation Factors during Cellular Stress and Cancer Progression.J Nucleic Acids. 2016;2016:8235121. doi: 10.1155/2016/8235121. Epub 2016 Dec 19. J Nucleic Acids. 2016. PMID: 28083147 Free PMC article. Review.
-
Noncanonical Translation Initiation in Eukaryotes.Cold Spring Harb Perspect Biol. 2019 Apr 1;11(4):a032672. doi: 10.1101/cshperspect.a032672. Cold Spring Harb Perspect Biol. 2019. PMID: 29959190 Free PMC article. Review.
-
More than just scanning: the importance of cap-independent mRNA translation initiation for cellular stress response and cancer.Cell Mol Life Sci. 2017 May;74(9):1659-1680. doi: 10.1007/s00018-016-2428-2. Epub 2016 Dec 2. Cell Mol Life Sci. 2017. PMID: 27913822 Free PMC article. Review.
-
The translational landscape as regulated by the RNA helicase DDX3.BMB Rep. 2022 Mar;55(3):125-135. doi: 10.5483/BMBRep.2022.55.3.188. BMB Rep. 2022. PMID: 35236544 Free PMC article. Review.
References
-
- O’Brien RJ, Wong PC. . Amyloid precursor protein processing and Alzheimer’s disease. Annu Rev Neurosci 2011; 34:185 - 204; http://dx.doi.org/10.1146/annurev-neuro-061010-113613; PMID: 21456963 - DOI - PMC - PubMed
-
- Zhang H, Ma Q, Zhang YW, Xu H. . Proteolytic processing of Alzheimer’s β-amyloid precursor protein. J Neurochem 2012; 120:Suppl 1 9 - 21; http://dx.doi.org/10.1111/j.1471-4159.2011.07519.x; PMID: 22122372 - DOI - PMC - PubMed
-
- Tam JH, Pasternak SH. . Amyloid and Alzheimer’s disease: inside and out. Can J Neurol Sci 2012; 39:286 - 98; PMID: 22547507 - PubMed
-
- Sun X, Bromley-Brits K, Song W. . Regulation of β-site APP-cleaving enzyme 1 gene expression and its role in Alzheimer’s disease. J Neurochem 2012; 120:Suppl 1 62 - 70; http://dx.doi.org/10.1111/j.1471-4159.2011.07515.x; PMID: 22122349 - DOI - PubMed
-
- Rogers GW Jr., Edelman GM, Mauro VP. . Differential utilization of upstream AUGs in the beta-secretase mRNA suggests that a shunting mechanism regulates translation. Proc Natl Acad Sci U S A 2004; 101:2794 - 9; http://dx.doi.org/10.1073/pnas.0308576101; PMID: 14981268 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
