Expression of the Retrotransposon Helena Reveals a Complex Pattern of TE Deregulation in Drosophila Hybrids
- PMID: 26812285
- PMCID: PMC4728067
- DOI: 10.1371/journal.pone.0147903
Expression of the Retrotransposon Helena Reveals a Complex Pattern of TE Deregulation in Drosophila Hybrids
Abstract
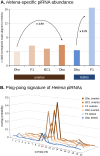
Transposable elements (TEs), repeated mobile sequences, are ubiquitous in the eukaryotic kingdom. Their mobilizing capacity confers on them a high mutagenic potential, which must be strongly regulated to guarantee genome stability. In the Drosophila germline, a small RNA-mediated silencing system, the piRNA (Piwi-interacting RNA) pathway, is the main responsible TE regulating mechanism, but some stressful conditions can destabilize it. For instance, during interspecific hybridization, genomic stress caused by the shock of two different genomes can lead, in both animals and plants, to higher transposition rates. A recent study in D. buzatii-D. koepferae hybrids detected mobilization of 28 TEs, yet little is known about the molecular mechanisms explaining this transposition release. We have characterized one of the mobilized TEs, the retrotransposon Helena, and used quantitative expression to assess whether its high transposition rates in hybrids are preceded by increased expression. We have also localized Helena expression in the gonads to see if cellular expression patterns have changed in the hybrids. To give more insight into changes in TE regulation in hybrids, we analysed Helena-specific piRNA populations of hybrids and parental species. Helena expression is not globally altered in somatic tissues, but male and female gonads have different patterns of deregulation. In testes, Helena is repressed in F1, increasing then its expression up to parental values. This is linked with a mislocation of Helena transcripts along with an increase of their specific piRNA levels. Ovaries have additive levels of Helena expression, but the ping-pong cycle efficiency seems to be reduced in F1 hybrids. This could be at the origin of new Helena insertions in hybrids, which would be transmitted to F1 hybrid female progeny.
Conflict of interest statement
Figures

Similar articles
-
Transposable Element Misregulation Is Linked to the Divergence between Parental piRNA Pathways in Drosophila Hybrids.Genome Biol Evol. 2017 Jun 1;9(6):1450-1470. doi: 10.1093/gbe/evx091. Genome Biol Evol. 2017. PMID: 28854624 Free PMC article.
-
Changes of Osvaldo expression patterns in germline of male hybrids between the species Drosophila buzzatii and Drosophila koepferae.Mol Genet Genomics. 2015 Aug;290(4):1471-83. doi: 10.1007/s00438-015-1012-z. Epub 2015 Feb 25. Mol Genet Genomics. 2015. PMID: 25711309
-
Transposable Element Expression and Regulation Profile in Gonads of Interspecific Hybrids of Drosophila arizonae and Drosophila mojavensis wrigleyi.Cells. 2021 Dec 18;10(12):3574. doi: 10.3390/cells10123574. Cells. 2021. PMID: 34944084 Free PMC article.
-
Silencing of Transposable Elements by piRNAs in Drosophila: An Evolutionary Perspective.Genomics Proteomics Bioinformatics. 2017 Jun;15(3):164-176. doi: 10.1016/j.gpb.2017.01.006. Epub 2017 Jun 8. Genomics Proteomics Bioinformatics. 2017. PMID: 28602845 Free PMC article. Review.
-
Small RNA Pathways That Protect the Somatic Genome.Int J Mol Sci. 2017 Apr 26;18(5):912. doi: 10.3390/ijms18050912. Int J Mol Sci. 2017. PMID: 28445427 Free PMC article. Review.
Cited by
-
High Stability of the Epigenome in Drosophila Interspecific Hybrids.Genome Biol Evol. 2022 Feb 4;14(2):evac024. doi: 10.1093/gbe/evac024. Genome Biol Evol. 2022. PMID: 35143649 Free PMC article.
-
Transposable Element Misregulation Is Linked to the Divergence between Parental piRNA Pathways in Drosophila Hybrids.Genome Biol Evol. 2017 Jun 1;9(6):1450-1470. doi: 10.1093/gbe/evx091. Genome Biol Evol. 2017. PMID: 28854624 Free PMC article.
-
Transposable Elements: Major Players in Shaping Genomic and Evolutionary Patterns.Cells. 2022 Mar 19;11(6):1048. doi: 10.3390/cells11061048. Cells. 2022. PMID: 35326499 Free PMC article. Review.
-
Assessing the Impact of a Viral Infection on the Expression of Transposable Elements in the Cabbage Looper Moth (Trichoplusia ni).Genome Biol Evol. 2021 Nov 5;13(11):evab231. doi: 10.1093/gbe/evab231. Genome Biol Evol. 2021. PMID: 34613390 Free PMC article.
-
Helena and BS: Two Travellers between the Genera Drosophila and Zaprionus.Genome Biol Evol. 2018 Oct 1;10(10):2671-2685. doi: 10.1093/gbe/evy184. Genome Biol Evol. 2018. PMID: 30165545 Free PMC article.
References
-
- Fontdevila A (2005) Hybrid genome evolution by transposition. Cytogenet Genome Res 110: 49–55. - PubMed
-
- Mallet J (2005) Hybridization as an invasion of the genome. Trends Ecol Evol 20: 229–237. - PubMed
-
- O’Neill RJW, O’Neill MJ, Marshall Graves JA (1998) Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393: 68–73. - PubMed
-
- Liu B, Wendel JF (2000) Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 43: 874–880. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

