Essentiality Assessment of Cysteinyl and Lysyl-tRNA Synthetases of Mycobacterium smegmatis
- PMID: 26794499
- PMCID: PMC4721953
- DOI: 10.1371/journal.pone.0147188
Essentiality Assessment of Cysteinyl and Lysyl-tRNA Synthetases of Mycobacterium smegmatis
Abstract
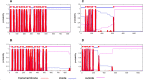
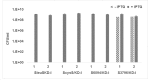
Discovery of mupirocin, an antibiotic that targets isoleucyl-tRNA synthetase, established aminoacyl-tRNA synthetase as an attractive target for the discovery of novel antibacterial agents. Despite a high degree of similarity between the bacterial and human aminoacyl-tRNA synthetases, the selectivity observed with mupirocin triggered the possibility of targeting other aminoacyl-tRNA synthetases as potential drug targets. These enzymes catalyse the condensation of a specific amino acid to its cognate tRNA in an energy-dependent reaction. Therefore, each organism is expected to encode at least twenty aminoacyl-tRNA synthetases, one for each amino acid. However, a bioinformatics search for genes encoding aminoacyl-tRNA synthetases from Mycobacterium smegmatis returned multiple genes for glutamyl (GluRS), cysteinyl (CysRS), prolyl (ProRS) and lysyl (LysRS) tRNA synthetases. The pathogenic mycobacteria, namely, Mycobacterium tuberculosis and Mycobacterium leprae, were also found to possess two genes each for CysRS and LysRS. A similar search indicated the presence of additional genes for LysRS in gram negative bacteria as well. Herein, we describe sequence and structural analysis of the additional aminoacyl-tRNA synthetase genes found in M. smegmatis. Characterization of conditional expression strains of Cysteinyl and Lysyl-tRNA synthetases generated in M. smegmatis revealed that the canonical aminoacyl-tRNA synthetase are essential, while the additional ones are not essential for the growth of M. smegmatis.
Conflict of interest statement
Figures
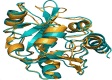


Similar articles
-
A bacterial ortholog of class II lysyl-tRNA synthetase activates lysine.FEBS Lett. 2010 Jul 16;584(14):3055-60. doi: 10.1016/j.febslet.2010.05.036. Epub 2010 May 24. FEBS Lett. 2010. PMID: 20580719 Free PMC article.
-
Aminoacyl-tRNA synthesis in Archaea.Nucleic Acids Symp Ser. 1997;(37):305-6. Nucleic Acids Symp Ser. 1997. PMID: 9586121
-
Functional association between three archaeal aminoacyl-tRNA synthetases.J Biol Chem. 2007 Feb 9;282(6):3680-7. doi: 10.1074/jbc.M609988200. Epub 2006 Dec 11. J Biol Chem. 2007. PMID: 17158871
-
Structural analyses of the malaria parasite aminoacyl-tRNA synthetases provide new avenues for antimalarial drug discovery.Protein Sci. 2021 Sep;30(9):1793-1803. doi: 10.1002/pro.4148. Protein Sci. 2021. PMID: 34184352 Free PMC article. Review.
-
Aminoacyl-tRNA synthesis: a postgenomic perspective.Cold Spring Harb Symp Quant Biol. 2001;66:175-83. doi: 10.1101/sqb.2001.66.175. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762020 Review. No abstract available.
Cited by
-
Diversification of aminoacyl-tRNA synthetase activities via genomic duplication.Front Physiol. 2022 Aug 19;13:983245. doi: 10.3389/fphys.2022.983245. eCollection 2022. Front Physiol. 2022. PMID: 36060688 Free PMC article. Review.
-
Antimicrobial peptides as drugs with double response against Mycobacterium tuberculosis coinfections in lung cancer.Front Microbiol. 2023 Jun 2;14:1183247. doi: 10.3389/fmicb.2023.1183247. eCollection 2023. Front Microbiol. 2023. PMID: 37342560 Free PMC article. Review.
-
Small RNA Profiling in Mycobacterium Provides Insights Into Stress Adaptability.Front Microbiol. 2021 Nov 4;12:752537. doi: 10.3389/fmicb.2021.752537. eCollection 2021. Front Microbiol. 2021. PMID: 34803973 Free PMC article.
-
Antimicrobial Peptides as Immunomodulators and Antimycobacterial Agents to Combat Mycobacterium tuberculosis: a Critical Review.Probiotics Antimicrob Proteins. 2023 Dec;15(6):1539-1566. doi: 10.1007/s12602-022-10018-6. Epub 2022 Dec 28. Probiotics Antimicrob Proteins. 2023. PMID: 36576687 Review.
-
Altered proteome of a Burkholderia pseudomallei mutant defective in short-chain dehydrogenase affects cell adhesion, biofilm formation and heat stress tolerance.PeerJ. 2020 Mar 19;8:e8659. doi: 10.7717/peerj.8659. eCollection 2020. PeerJ. 2020. PMID: 32219018 Free PMC article.
References
-
- Parenti MA, Hatfield SM, Leyden JJ. Mupirocin: a topical antibiotic with a unique structure and mechanism of action. Clin. Pharm. 1987, 6:761–770. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

