Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes
- PMID: 26787893
- PMCID: PMC4747710
- DOI: 10.1073/pnas.1518280113
Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes
Abstract
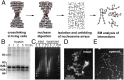
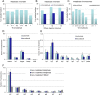
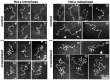
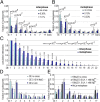
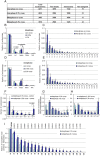
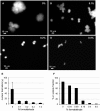
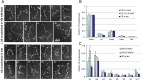
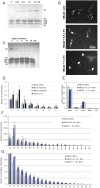
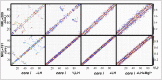
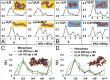
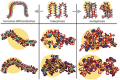
The architecture of higher-order chromatin in eukaryotic cell nuclei is largely unknown. Here, we use electron microscopy-assisted nucleosome interaction capture (EMANIC) cross-linking experiments in combination with mesoscale chromatin modeling of 96-nucleosome arrays to investigate the internal organization of condensed chromatin in interphase cell nuclei and metaphase chromosomes at nucleosomal resolution. The combined data suggest a novel hierarchical looping model for chromatin higher-order folding, similar to rope flaking used in mountain climbing and rappelling. Not only does such packing help to avoid tangling and self-crossing, it also facilitates rope unraveling. Hierarchical looping is characterized by an increased frequency of higher-order internucleosome contacts for metaphase chromosomes compared with chromatin fibers in vitro and interphase chromatin, with preservation of a dominant two-start zigzag organization associated with the 30-nm fiber. Moreover, the strong dependence of looping on linker histone concentration suggests a hierarchical self-association mechanism of relaxed nucleosome zigzag chains rather than longitudinal compaction as seen in 30-nm fibers. Specifically, concentrations lower than one linker histone per nucleosome promote self-associations and formation of these looped networks of zigzag fibers. The combined experimental and modeling evidence for condensed metaphase chromatin as hierarchical loops and bundles of relaxed zigzag nucleosomal chains rather than randomly coiled threads or straight and stiff helical fibers reconciles aspects of other models for higher-order chromatin structure; it constitutes not only an efficient storage form for the genomic material, consistent with other genome-wide chromosome conformation studies that emphasize looping, but also a convenient organization for local DNA unraveling and genome access.
Keywords: chromatin higher-order structure; electron microscopy; linker histone; mesoscale modeling; nucleosome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Linking Chromatin Fibers to Gene Folding by Hierarchical Looping.Biophys J. 2017 Feb 7;112(3):434-445. doi: 10.1016/j.bpj.2017.01.003. Epub 2017 Jan 31. Biophys J. 2017. PMID: 28153411 Free PMC article. Review.
-
Electron microscopy and atomic force microscopy studies of chromatin and metaphase chromosome structure.Micron. 2011 Dec;42(8):733-50. doi: 10.1016/j.micron.2011.05.002. Epub 2011 May 12. Micron. 2011. PMID: 21703860 Review.
-
Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions.Proc Natl Acad Sci U S A. 2009 Aug 11;106(32):13317-22. doi: 10.1073/pnas.0903280106. Epub 2009 Jul 27. Proc Natl Acad Sci U S A. 2009. PMID: 19651606 Free PMC article.
-
Mesoscale Modeling Reveals Hierarchical Looping of Chromatin Fibers Near Gene Regulatory Elements.J Phys Chem B. 2016 Aug 25;120(33):8642-53. doi: 10.1021/acs.jpcb.6b03197. Epub 2016 Jun 16. J Phys Chem B. 2016. PMID: 27218881 Free PMC article.
-
Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin.Proc Natl Acad Sci U S A. 1998 Nov 24;95(24):14173-8. doi: 10.1073/pnas.95.24.14173. Proc Natl Acad Sci U S A. 1998. PMID: 9826673 Free PMC article.
Cited by
-
Hypothesis: The opposing pulling forces exerted by spindle microtubules can cause sliding of chromatin layers and facilitate sister chromatid resolution.Front Genet. 2023 Nov 24;14:1321260. doi: 10.3389/fgene.2023.1321260. eCollection 2023. Front Genet. 2023. PMID: 38075677 Free PMC article.
-
Budding yeast chromatin is dispersed in a crowded nucleoplasm in vivo.Mol Biol Cell. 2016 Nov 1;27(21):3357-3368. doi: 10.1091/mbc.E16-07-0506. Epub 2016 Sep 7. Mol Biol Cell. 2016. PMID: 27605704 Free PMC article.
-
Nucleosome spacing periodically modulates nucleosome chain folding and DNA topology in circular nucleosome arrays.J Biol Chem. 2019 Mar 15;294(11):4233-4246. doi: 10.1074/jbc.RA118.006412. Epub 2019 Jan 10. J Biol Chem. 2019. PMID: 30630950 Free PMC article.
-
Large-scale simulations of nucleoprotein complexes: ribosomes, nucleosomes, chromatin, chromosomes and CRISPR.Curr Opin Struct Biol. 2019 Apr;55:104-113. doi: 10.1016/j.sbi.2019.03.004. Epub 2019 May 21. Curr Opin Struct Biol. 2019. PMID: 31125796 Free PMC article. Review.
-
HMGN1 and 2 remodel core and linker histone tail domains within chromatin.Nucleic Acids Res. 2017 Sep 29;45(17):9917-9930. doi: 10.1093/nar/gkx579. Nucleic Acids Res. 2017. PMID: 28973435 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

