Epigenetic Therapeutics: A New Weapon in the War Against Cancer
- PMID: 26768237
- PMCID: PMC4937439
- DOI: 10.1146/annurev-med-111314-035900
Epigenetic Therapeutics: A New Weapon in the War Against Cancer
Abstract
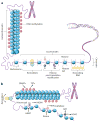
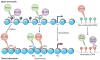
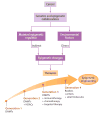
The past 15 years have seen an explosion of discoveries related to the cellular regulation of phenotypes through epigenetic mechanisms. This regulation provides a software that packages DNA, without changing the primary base sequence, to establish heritable patterns of gene expression. In cancer, many aspects of the epigenome, controlled by DNA methylation, chromatin, and nucleosome positioning, are altered as one means by which tumor cells maintain abnormal states of self-renewal at the expense of normal maturation. Epigenetic and genetic abnormalities thus collaborate in cancer initiation and progression, as exemplified by frequent mutations in genes encoding proteins that control the epigenome. There is growing emphasis on using epigenetic therapies to reprogram neoplastic cells toward a normal state. Many agents targeting epigenetic regulation are under development and entering clinical trials. This review highlights the promise that epigenetic therapy, often in combination with other therapies, will become a potent tool for cancer management over the next decade.
Keywords: DNA methyltransferase inhibitor; epigenetics; histone deacetylase inhibitor therapy.
Figures
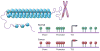
Similar articles
-
Epigenetic therapy for solid tumors: from bench science to clinical trials.Epigenomics. 2015;7(2):215-35. doi: 10.2217/epi.14.73. Epigenomics. 2015. PMID: 25942532 Review.
-
Human DNA (cytosine-5)-methyltransferases: a functional and structural perspective for epigenetic cancer therapy.Biochimie. 2017 Aug;139:137-147. doi: 10.1016/j.biochi.2017.06.003. Epub 2017 Jun 6. Biochimie. 2017. PMID: 28600135 Review.
-
Epigenetic therapy of cancer with histone deacetylase inhibitors.J Cancer Res Ther. 2014 Jul-Sep;10(3):469-78. doi: 10.4103/0973-1482.137937. J Cancer Res Ther. 2014. PMID: 25313724 Review.
-
Epigenetic therapy and chemosensitization in solid malignancy.Cancer Treat Rev. 2017 Apr;55:200-208. doi: 10.1016/j.ctrv.2017.03.008. Epub 2017 Apr 8. Cancer Treat Rev. 2017. PMID: 28431263 Review.
-
Epigenomes as therapeutic targets.Pharmacol Ther. 2015 Jul;151:72-86. doi: 10.1016/j.pharmthera.2015.03.003. Epub 2015 Mar 20. Pharmacol Ther. 2015. PMID: 25797698 Review.
Cited by
-
A multiomics approach reveals RNA dynamics promote cellular sensitivity to DNA hypomethylation.Sci Rep. 2024 Oct 29;14(1):25940. doi: 10.1038/s41598-024-77314-9. Sci Rep. 2024. PMID: 39472491 Free PMC article.
-
Comprehensive review and updated analysis of DNA methylation in hepatocellular carcinoma: From basic research to clinical application.Clin Transl Med. 2024 Nov;14(11):e70066. doi: 10.1002/ctm2.70066. Clin Transl Med. 2024. PMID: 39462685 Free PMC article. Review.
-
Targeting the cancer epigenome for therapy.Nat Rev Genet. 2016 Sep 15;17(10):630-41. doi: 10.1038/nrg.2016.93. Nat Rev Genet. 2016. PMID: 27629931 Review.
-
Nucleosome conformation dictates the histone code.Elife. 2024 Feb 6;13:e78866. doi: 10.7554/eLife.78866. Elife. 2024. PMID: 38319148 Free PMC article.
-
Insulin and Metformin Control Cell Proliferation by Regulating TDG-Mediated DNA Demethylation in Liver and Breast Cancer Cells.Mol Ther Oncolytics. 2020 Jun 24;18:282-294. doi: 10.1016/j.omto.2020.06.010. eCollection 2020 Sep 25. Mol Ther Oncolytics. 2020. PMID: 32728616 Free PMC article.
References
-
- Masters GA, Krilov L, Bailey HH, et al. Clinical cancer advances 2015: annual report on progress against cancer from the American Society of Clinical Oncology. J Clin Oncol. 2015;33:786–809. - PubMed
-
- Mwenifumbo JC, Marra MA. Cancer genome-sequencing study design. Nat Rev Genet. 2013;14:321–32. - PubMed
-
- Allis C, Jenuwein T, Reinberg D, Caparros M. Epigenetics. 2 Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2015.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

