Recognition of the nucleosome by chromatin factors and enzymes
- PMID: 26764865
- PMCID: PMC4834262
- DOI: 10.1016/j.sbi.2015.11.014
Recognition of the nucleosome by chromatin factors and enzymes
Abstract
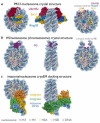
Dynamic expression of the genome requires coordinated binding of chromatin factors and enzymes that carry out genome-templated processes. Until recently, the molecular mechanisms governing how these factors and enzymes recognize and act on the fundamental unit of chromatin, the nucleosome core particle, have remained a mystery. A small, yet growing set of structures of the nucleosome in complex with chromatin factors and enzymes highlights the importance of multivalency in defining nucleosome binding and specificity. Many such interactions include an arginine anchor motif, which targets a unique acidic patch on the nucleosome surface. These emerging paradigms for chromatin recognition will be discussed, focusing on several recent structural breakthroughs.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
The nucleosome: orchestrating DNA damage signaling and repair within chromatin.Biochem Cell Biol. 2016 Oct;94(5):381-395. doi: 10.1139/bcb-2016-0017. Epub 2016 Apr 13. Biochem Cell Biol. 2016. PMID: 27240007 Review.
-
Comprehensive nucleosome interactome screen establishes fundamental principles of nucleosome binding.Nucleic Acids Res. 2020 Sep 25;48(17):9415-9432. doi: 10.1093/nar/gkaa544. Nucleic Acids Res. 2020. PMID: 32658293 Free PMC article.
-
Principles of nucleosome recognition by chromatin factors and enzymes.Curr Opin Struct Biol. 2021 Dec;71:16-26. doi: 10.1016/j.sbi.2021.05.006. Epub 2021 Jun 28. Curr Opin Struct Biol. 2021. PMID: 34198054 Free PMC article. Review.
-
Structural Basis of Nucleosome Recognition and Modulation.Bioessays. 2020 Sep;42(9):e1900234. doi: 10.1002/bies.201900234. Epub 2020 Jun 22. Bioessays. 2020. PMID: 32567715 Review.
-
Strategies for crystallizing a chromatin protein in complex with the nucleosome core particle.Anal Biochem. 2013 Nov 15;442(2):138-45. doi: 10.1016/j.ab.2013.07.038. Epub 2013 Aug 6. Anal Biochem. 2013. PMID: 23928047 Free PMC article.
Cited by
-
Deciphering principles of nucleosome interactions and impact of cancer-associated mutations from comprehensive interaction network analysis.Brief Bioinform. 2024 Jan 22;25(2):bbad532. doi: 10.1093/bib/bbad532. Brief Bioinform. 2024. PMID: 38329268 Free PMC article.
-
Characterization of nucleosome sediments for protein interaction studies by solid-state NMR spectroscopy.Magn Reson (Gott). 2021;2(1):187-202. doi: 10.5194/mr-2-187-2021. Epub 2021 Apr 21. Magn Reson (Gott). 2021. PMID: 35647606 Free PMC article.
-
Structural basis of specific H2A K13/K15 ubiquitination by RNF168.Nat Commun. 2019 Apr 15;10(1):1751. doi: 10.1038/s41467-019-09756-z. Nat Commun. 2019. PMID: 30988309 Free PMC article.
-
The SAGA continues: The rise of cis- and trans-histone crosstalk pathways.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194600. doi: 10.1016/j.bbagrm.2020.194600. Epub 2020 Jul 6. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32645359 Free PMC article. Review.
-
3.9 Å structure of the nucleosome core particle determined by phase-plate cryo-EM.Nucleic Acids Res. 2016 Sep 30;44(17):8013-9. doi: 10.1093/nar/gkw708. Epub 2016 Aug 25. Nucleic Acids Res. 2016. PMID: 27563056 Free PMC article.
References
-
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. - PubMed
-
- This is a classic paper reporting the first high-resolution structure of nucleosome core particle.
-
- Ng MK, Cheung P. A brief histone in time: understanding the combinatorial functions of histone PTMs in the nucleosome context. Biochem. Cell Biol. 2015 doi:10.1139/bcb-2015-0031. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

