Sampling From the Proteome to the Human Leukocyte Antigen-DR (HLA-DR) Ligandome Proceeds Via High Specificity
- PMID: 26764012
- PMCID: PMC4824864
- DOI: 10.1074/mcp.M115.055780
Sampling From the Proteome to the Human Leukocyte Antigen-DR (HLA-DR) Ligandome Proceeds Via High Specificity
Abstract
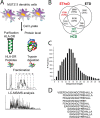
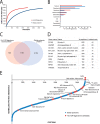
Comprehensive analysis of the complex nature of the Human Leukocyte Antigen (HLA) class II ligandome is of utmost importance to understand the basis for CD4(+)T cell mediated immunity and tolerance. Here, we implemented important improvements in the analysis of the repertoire of HLA-DR-presented peptides, using hybrid mass spectrometry-based peptide fragmentation techniques on a ligandome sample isolated from matured human monocyte-derived dendritic cells (DC). The reported data set constitutes nearly 14 thousand unique high-confident peptides,i.e.the largest single inventory of human DC derived HLA-DR ligands to date. From a technical viewpoint the most prominent finding is that no single peptide fragmentation technique could elucidate the majority of HLA-DR ligands, because of the wide range of physical chemical properties displayed by the HLA-DR ligandome. Our in-depth profiling allowed us to reveal a strikingly poor correlation between the source proteins identified in the HLA class II ligandome and the DC cellular proteome. Important selective sieving from the sampled proteome to the ligandome was evidenced by specificity in the sequences of the core regions both at their N- and C- termini, hence not only reflecting binding motifs but also dominant protease activity associated to the endolysosomal compartments. Moreover, we demonstrate that the HLA-DR ligandome reflects a surface representation of cell-compartments specific for biological events linked to the maturation of monocytes into antigen presenting cells. Our results present new perspectives into the complex nature of the HLA class II system and will aid future immunological studies in characterizing the full breadth of potential CD4(+)T cell epitopes relevant in health and disease.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Comparative profiling of HLA-DR and HLA-DQ associated factor VIII peptides presented by monocyte-derived dendritic cells.Haematologica. 2018 Jan;103(1):172-178. doi: 10.3324/haematol.2017.175083. Epub 2017 Oct 12. Haematologica. 2018. PMID: 29025906 Free PMC article.
-
Ligandomes obtained from different HLA-class II-molecules are homologous for N- and C-terminal residues outside the peptide-binding cleft.Immunogenetics. 2019 Sep;71(8-9):519-530. doi: 10.1007/s00251-019-01129-6. Epub 2019 Sep 13. Immunogenetics. 2019. PMID: 31520135 Free PMC article.
-
The human leukocyte antigen-presented ligandome of B lymphocytes.Mol Cell Proteomics. 2013 Jul;12(7):1829-43. doi: 10.1074/mcp.M112.024810. Epub 2013 Mar 12. Mol Cell Proteomics. 2013. PMID: 23481700 Free PMC article.
-
Mapping the tumour human leukocyte antigen (HLA) ligandome by mass spectrometry.Immunology. 2018 Jul;154(3):331-345. doi: 10.1111/imm.12936. Epub 2018 May 8. Immunology. 2018. PMID: 29658117 Free PMC article. Review.
-
What Is the Role of HLA-I on Cancer Derived Extracellular Vesicles? Defining the Challenges in Characterisation and Potential Uses of This Ligandome.Int J Mol Sci. 2021 Dec 17;22(24):13554. doi: 10.3390/ijms222413554. Int J Mol Sci. 2021. PMID: 34948350 Free PMC article. Review.
Cited by
-
Determining the Quantitative Principles of T Cell Response to Antigenic Disparity in Stem Cell Transplantation.Front Immunol. 2018 Oct 11;9:2284. doi: 10.3389/fimmu.2018.02284. eCollection 2018. Front Immunol. 2018. PMID: 30364159 Free PMC article. Review.
-
Computational characterization of the peptidome in transporter associated with antigen processing (TAP)-deficient cells.PLoS One. 2019 Jan 15;14(1):e0210583. doi: 10.1371/journal.pone.0210583. eCollection 2019. PLoS One. 2019. PMID: 30645615 Free PMC article.
-
Comparative profiling of HLA-DR and HLA-DQ associated factor VIII peptides presented by monocyte-derived dendritic cells.Haematologica. 2018 Jan;103(1):172-178. doi: 10.3324/haematol.2017.175083. Epub 2017 Oct 12. Haematologica. 2018. PMID: 29025906 Free PMC article.
-
Reducing Immunogenicity by Design: Approaches to Minimize Immunogenicity of Monoclonal Antibodies.BioDrugs. 2024 Mar;38(2):205-226. doi: 10.1007/s40259-023-00641-2. Epub 2024 Jan 23. BioDrugs. 2024. PMID: 38261155 Free PMC article. Review.
-
HLA Class II Presentation Is Specifically Altered at Elevated Temperatures in the B-Lymphoblastic Cell Line JY.Mol Cell Proteomics. 2021;20:100089. doi: 10.1016/j.mcpro.2021.100089. Epub 2021 Apr 29. Mol Cell Proteomics. 2021. PMID: 33933681 Free PMC article.
References
-
- Neefjes J., Jongsma M. L., Paul P., and Bakke O. (2011) Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 - PubMed
-
- Adamopoulou E., Tenzer S., Hillen N., Klug P., Rota I. a, Tietz S., Gebhardt M., Stevanovic S., Schild H., Tolosa E., Melms A., and Stoeckle C. (2013) Exploring the MHC-peptide matrix of central tolerance in the human thymus. Nat. Commun. 4, 2039. - PubMed
-
- Suri A., Lovitch S. B., and Unanue E. R. (2006) The wide diversity and complexity of peptides bound to class II MHC molecules. Curr. Opin. Immunol. 18, 70–77 - PubMed
-
- Ovsyannikova I. G., Johnson K. L., Bergen H. R., and Poland G. A. (2007) Mass spectrometry and peptide-based vaccine development. Clin. Pharmacol. Ther. 82, 644–652 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

