Open Source Bayesian Models. 3. Composite Models for Prediction of Binned Responses
- PMID: 26750305
- PMCID: PMC4764945
- DOI: 10.1021/acs.jcim.5b00555
Open Source Bayesian Models. 3. Composite Models for Prediction of Binned Responses
Abstract
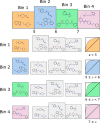
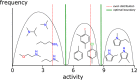
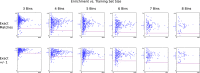
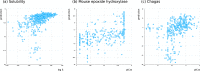
Bayesian models constructed from structure-derived fingerprints have been a popular and useful method for drug discovery research when applied to bioactivity measurements that can be effectively classified as active or inactive. The results can be used to rank candidate structures according to their probability of activity, and this ranking benefits from the high degree of interpretability when structure-based fingerprints are used, making the results chemically intuitive. Besides selecting an activity threshold, building a Bayesian model is fast and requires few or no parameters or user intervention. The method also does not suffer from such acute overtraining problems as quantitative structure-activity relationships or quantitative structure-property relationships (QSAR/QSPR). This makes it an approach highly suitable for automated workflows that are independent of user expertise or prior knowledge of the training data. We now describe a new method for creating a composite group of Bayesian models to extend the method to work with multiple states, rather than just binary. Incoming activities are divided into bins, each covering a mutually exclusive range of activities. For each of these bins, a Bayesian model is created to model whether or not the compound belongs in the bin. Analyzing putative molecules using the composite model involves making a prediction for each bin and examining the relative likelihood for each assignment, for example, highest value wins. The method has been evaluated on a collection of hundreds of data sets extracted from ChEMBL v20 and validated data sets for ADME/Tox and bioactivity.
Conflict of interest statement
The authors declare the following competing financial interests: S.E. is a consultant for Collaborative Drug Discovery, Inc. A.M.C. is the founder of Molecular Materials Informatics, Inc.
Figures



Similar articles
-
Open Source Bayesian Models. 1. Application to ADME/Tox and Drug Discovery Datasets.J Chem Inf Model. 2015 Jun 22;55(6):1231-45. doi: 10.1021/acs.jcim.5b00143. Epub 2015 Jun 3. J Chem Inf Model. 2015. PMID: 25994950 Free PMC article.
-
Introduction of a methodology for visualization and graphical interpretation of Bayesian classification models.J Chem Inf Model. 2014 Sep 22;54(9):2451-68. doi: 10.1021/ci500410g. Epub 2014 Aug 27. J Chem Inf Model. 2014. PMID: 25137527
-
Profile-QSAR: a novel meta-QSAR method that combines activities across the kinase family to accurately predict affinity, selectivity, and cellular activity.J Chem Inf Model. 2011 Aug 22;51(8):1942-56. doi: 10.1021/ci1005004. Epub 2011 Jul 19. J Chem Inf Model. 2011. PMID: 21667971
-
[Ring-system-based Chemical Structure Enumeration for de Novo Design].Yakugaku Zasshi. 2016;136(1):101-6. doi: 10.1248/yakushi.15-00230-2. Yakugaku Zasshi. 2016. PMID: 26725676 Review. Japanese.
-
Applying machine learning techniques for ADME-Tox prediction: a review.Expert Opin Drug Metab Toxicol. 2015 Feb;11(2):259-71. doi: 10.1517/17425255.2015.980814. Epub 2014 Dec 2. Expert Opin Drug Metab Toxicol. 2015. PMID: 25440524 Review.
Cited by
-
The Next Era: Deep Learning in Pharmaceutical Research.Pharm Res. 2016 Nov;33(11):2594-603. doi: 10.1007/s11095-016-2029-7. Epub 2016 Sep 6. Pharm Res. 2016. PMID: 27599991 Free PMC article. Review.
-
Machine Learning Platform to Discover Novel Growth Inhibitors of Neisseria gonorrhoeae.Pharm Res. 2020 Jul 13;37(7):141. doi: 10.1007/s11095-020-02876-y. Pharm Res. 2020. PMID: 32661900 Free PMC article.
-
Machine Learning Model Analysis and Data Visualization with Small Molecules Tested in a Mouse Model of Mycobacterium tuberculosis Infection (2014-2015).J Chem Inf Model. 2016 Jul 25;56(7):1332-43. doi: 10.1021/acs.jcim.6b00004. Epub 2016 Jul 1. J Chem Inf Model. 2016. PMID: 27335215 Free PMC article.
-
Comparing Multiple Machine Learning Algorithms and Metrics for Estrogen Receptor Binding Prediction.Mol Pharm. 2018 Oct 1;15(10):4361-4370. doi: 10.1021/acs.molpharmaceut.8b00546. Epub 2018 Aug 28. Mol Pharm. 2018. PMID: 30114914 Free PMC article.
-
Data Mining and Computational Modeling of High-Throughput Screening Datasets.Methods Mol Biol. 2018;1755:197-221. doi: 10.1007/978-1-4939-7724-6_14. Methods Mol Biol. 2018. PMID: 29671272 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

