The mysterious orphans of Mycoplasmataceae
- PMID: 26747447
- PMCID: PMC4706650
- DOI: 10.1186/s13062-015-0104-3
The mysterious orphans of Mycoplasmataceae
Abstract
Background: The length of a protein sequence is largely determined by its function. In certain species, it may be also affected by additional factors, such as growth temperature or acidity. In 2002, it was shown that in the bacterium Escherichia coli and in the archaeon Archaeoglobus fulgidus, protein sequences with no homologs were, on average, shorter than those with homologs (BMC Evol Biol 2:20, 2002). It is now generally accepted that in bacterial and archaeal genomes the distributions of protein length are different between sequences with and without homologs. In this study, we examine this postulate by conducting a comprehensive analysis of all annotated prokaryotic genomes and by focusing on certain exceptions.
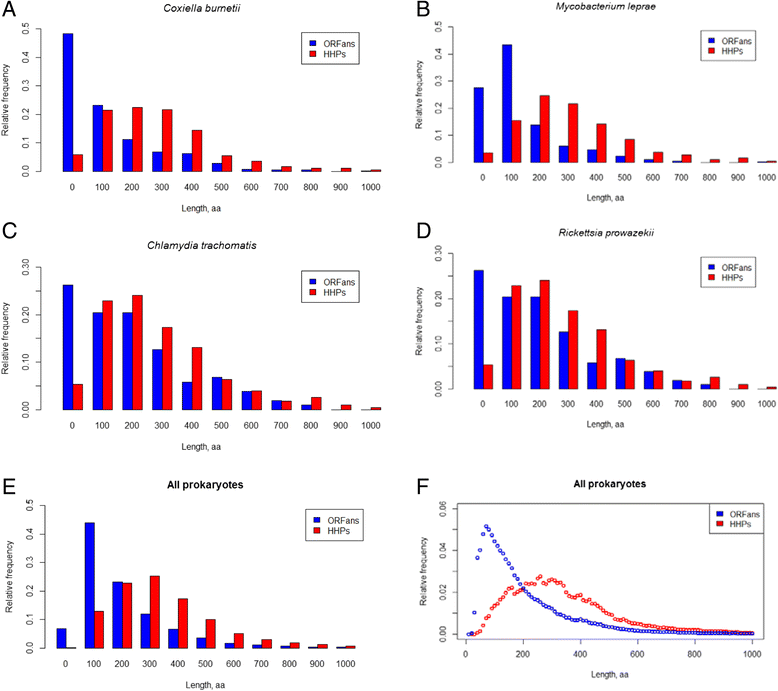
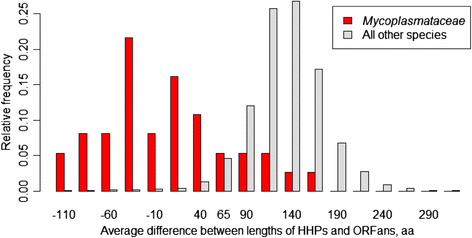
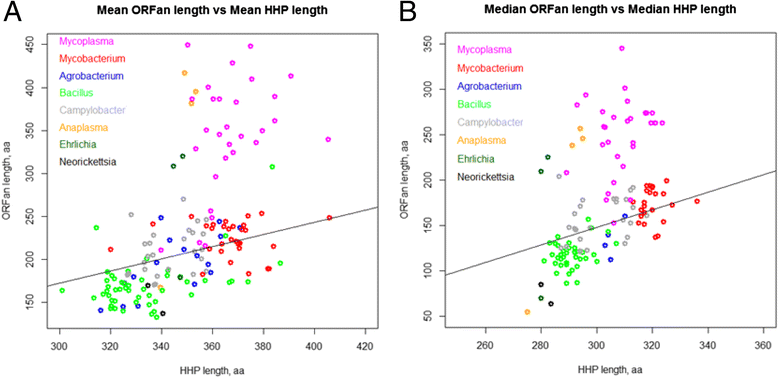
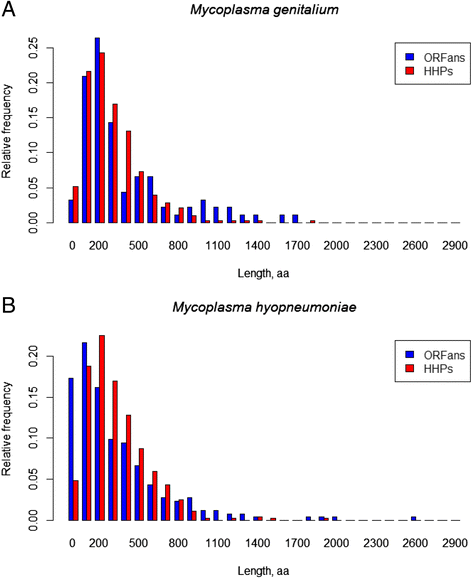
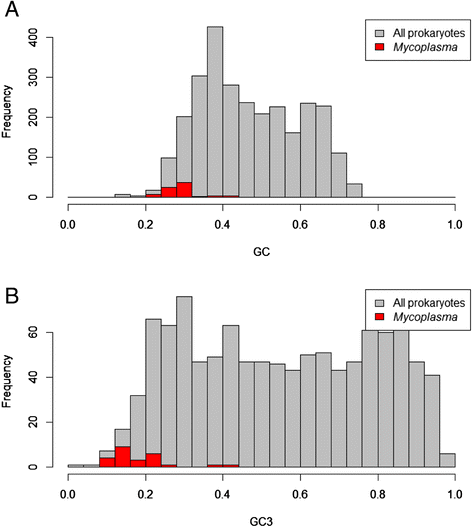
Results: We compared the distribution of lengths of "having homologs proteins" (HHPs) and "non-having homologs proteins" (orphans or ORFans) in all currently completely sequenced and COG-annotated prokaryotic genomes. As expected, the HHPs and ORFans have strikingly different length distributions in almost all genomes. As previously established, the HHPs, indeed are, on average, longer than the ORFans, and the length distributions for the ORFans have a relatively narrow peak, in contrast to the HHPs, whose lengths spread over a wider range of values. However, about thirty genomes do not obey these rules. Practically all genomes of Mycoplasma and Ureaplasma have atypical ORFans distributions, with the mean lengths of ORFan larger than the mean lengths of HHPs. These genera constitute over 80 % of atypical genomes.
Conclusions: We confirmed on a ubiquitous set of genomes that the previous observation of HHPs and ORFans have different gene length distributions. We also showed that Mycoplasmataceae genomes have very distinctive distributions of ORFans lengths. We offer several possible biological explanations of this phenomenon, such as an adaptation to Mycoplasmataceae's ecological niche, specifically its "quiet" co-existence with host organisms, resulting in long ABC transporters.
Figures
Similar articles
-
Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli.Genome Res. 2004 Jun;14(6):1036-42. doi: 10.1101/gr.2231904. Genome Res. 2004. PMID: 15173110 Free PMC article.
-
Population diversity of ORFan genes in Escherichia coli.Genome Biol Evol. 2012;4(11):1176-87. doi: 10.1093/gbe/evs081. Genome Biol Evol. 2012. PMID: 23034216 Free PMC article.
-
Identification and investigation of ORFans in the viral world.BMC Genomics. 2008 Jan 19;9:24. doi: 10.1186/1471-2164-9-24. BMC Genomics. 2008. PMID: 18205946 Free PMC article.
-
Analysis of singleton ORFans in fully sequenced microbial genomes.Proteins. 2003 Nov 1;53(2):241-51. doi: 10.1002/prot.10423. Proteins. 2003. PMID: 14517975
-
Twenty thousand ORFan microbial protein families for the biologist?Structure. 2003 Jan;11(1):7-9. doi: 10.1016/s0969-2126(02)00938-3. Structure. 2003. PMID: 12517334 Review.
Cited by
-
Nucleotide diversity analysis highlights functionally important genomic regions.Sci Rep. 2016 Oct 24;6:35730. doi: 10.1038/srep35730. Sci Rep. 2016. PMID: 27774999 Free PMC article.
-
Discovery of numerous novel small genes in the intergenic regions of the Escherichia coli O157:H7 Sakai genome.PLoS One. 2017 Sep 13;12(9):e0184119. doi: 10.1371/journal.pone.0184119. eCollection 2017. PLoS One. 2017. PMID: 28902868 Free PMC article.
-
Nasopulmonary mites (Acari: Halarachnidae) as potential vectors of bacterial pathogens, including Streptococcus phocae, in marine mammals.PLoS One. 2022 Jun 16;17(6):e0270009. doi: 10.1371/journal.pone.0270009. eCollection 2022. PLoS One. 2022. PMID: 35709209 Free PMC article.
-
Comparative Analysis of Mycoplasma gallisepticum vlhA Promoters.Front Genet. 2018 Nov 21;9:569. doi: 10.3389/fgene.2018.00569. eCollection 2018. Front Genet. 2018. PMID: 30519256 Free PMC article.
-
No Evidence for Phylostratigraphic Bias Impacting Inferences on Patterns of Gene Emergence and Evolution.Mol Biol Evol. 2017 Apr 1;34(4):843-856. doi: 10.1093/molbev/msw284. Mol Biol Evol. 2017. PMID: 28087778 Free PMC article.
References
-
- Galperin MY, Tatusov RL, Koonin EV. Comparing microbila genomes: how the gene set determines the lifestyle. In: Charlebois RL, editor. Organization of the prokaryotic genome. Washington, DC: ASM Press; 1999.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

