A basic helix-loop-helix transcription factor, PhFBH4, regulates flower senescence by modulating ethylene biosynthesis pathway in petunia
- PMID: 26715989
- PMCID: PMC4680862
- DOI: 10.1038/hortres.2015.59
A basic helix-loop-helix transcription factor, PhFBH4, regulates flower senescence by modulating ethylene biosynthesis pathway in petunia
Abstract
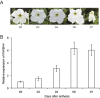
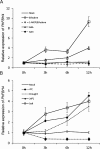
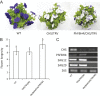
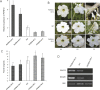
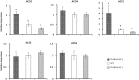
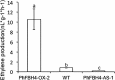
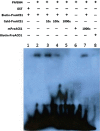
The basic helix-loop-helix (bHLH) transcription factors (TFs) play important roles in regulating multiple biological processes in plants. However, there are few reports about the function of bHLHs in flower senescence. In this study, a bHLH TF, PhFBH4, was found to be dramatically upregulated during flower senescence. Transcription of PhFBH4 is induced by plant hormones and abiotic stress treatments. Silencing of PhFBH4 using virus-induced gene silencing or an antisense approach extended flower longevity, while transgenic petunia flowers with an overexpression construct showed a reduction in flower lifespan. Abundance of transcripts of senescence-related genes (SAG12, SAG29) was significantly changed in petunia PhFBH4 transgenic flowers. Furthermore, silencing or overexpression of PhFBH4 reduced or increased, respectively, transcript abundances of important ethylene biosynthesis-related genes, ACS1 and ACO1, thereby influencing ethylene production. An electrophoretic mobility shift assay showed that the PhFBH4 protein physically interacted with the G-box cis-element in the promoter of ACS1, suggesting that ACS1 was a direct target of the PhFBH4 protein. In addition, ectopic expression of this gene altered plant development including plant height, internode length, and size of leaves and flowers, accompanied by alteration of transcript abundance of the gibberellin biosynthesis-related gene GA2OX3. Our results indicate that PhFBH4 plays an important role in regulating plant growth and development through modulating the ethylene biosynthesis pathway.
Figures

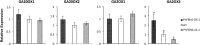
Similar articles
-
Molecular aspects of flower senescence and strategies to improve flower longevity.Breed Sci. 2018 Jan;68(1):99-108. doi: 10.1270/jsbbs.17081. Epub 2018 Feb 27. Breed Sci. 2018. PMID: 29681752 Free PMC article. Review.
-
A Petunia homeodomain-leucine zipper protein, PhHD-Zip, plays an important role in flower senescence.PLoS One. 2014 Feb 14;9(2):e88320. doi: 10.1371/journal.pone.0088320. eCollection 2014. PLoS One. 2014. PMID: 24551088 Free PMC article.
-
A petunia transcription factor, PhOBF1, regulates flower senescence by modulating gibberellin biosynthesis.Hortic Res. 2023 Feb 16;10(4):uhad022. doi: 10.1093/hr/uhad022. eCollection 2023 Apr. Hortic Res. 2023. PMID: 37786859 Free PMC article.
-
Overproduction of cytokinins in petunia flowers transformed with P(SAG12)-IPT delays corolla senescence and decreases sensitivity to ethylene.Plant Physiol. 2003 Aug;132(4):2174-83. doi: 10.1104/pp.103.023945. Plant Physiol. 2003. PMID: 12913172 Free PMC article.
-
Regulation of volatile benzenoid biosynthesis in petunia flowers.Trends Plant Sci. 2006 Jan;11(1):20-5. doi: 10.1016/j.tplants.2005.09.009. Epub 2005 Oct 12. Trends Plant Sci. 2006. PMID: 16226052 Review.
Cited by
-
Transcriptome Profiling of Petal Abscission Zone and Functional Analysis of an Aux/IAA Family Gene RhIAA16 Involved in Petal Shedding in Rose.Front Plant Sci. 2016 Sep 15;7:1375. doi: 10.3389/fpls.2016.01375. eCollection 2016. Front Plant Sci. 2016. PMID: 27695465 Free PMC article.
-
Molecular aspects of flower senescence and strategies to improve flower longevity.Breed Sci. 2018 Jan;68(1):99-108. doi: 10.1270/jsbbs.17081. Epub 2018 Feb 27. Breed Sci. 2018. PMID: 29681752 Free PMC article. Review.
-
The FBH family of bHLH transcription factors controls ACC synthase expression in sugarcane.J Exp Bot. 2018 Apr 27;69(10):2511-2525. doi: 10.1093/jxb/ery083. J Exp Bot. 2018. PMID: 29514290 Free PMC article.
-
LreEF1A4, a Translation Elongation Factor from Lilium regale, Is Pivotal for Cucumber Mosaic Virus and Tobacco Rattle Virus Infections and Tolerance to Salt and Drought.Int J Mol Sci. 2020 Mar 18;21(6):2083. doi: 10.3390/ijms21062083. Int J Mol Sci. 2020. PMID: 32197393 Free PMC article.
-
Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones.Front Plant Sci. 2017 Apr 4;8:475. doi: 10.3389/fpls.2017.00475. eCollection 2017. Front Plant Sci. 2017. PMID: 28421102 Free PMC article. Review.
References
-
- Wagstaff C, Yang TJW, Stead AD, Buchanan-Wollaston V, Roberts JA. A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. Plant J 2009; 57: 690–705. - PubMed
-
- Liu M, Diretto G, Pirrello J et al. The chimeric repressor version of an ethylene response factor (ERF) family member, Sl-ERF.B3, shows contrasting effects on tomato fruit ripening. New Phytol 2014; 203: 206–218. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

