8-Hydroxyquinoline-based inhibitors of the Rce1 protease disrupt Ras membrane localization in human cells
- PMID: 26706114
- PMCID: PMC4749511
- DOI: 10.1016/j.bmc.2015.11.043
8-Hydroxyquinoline-based inhibitors of the Rce1 protease disrupt Ras membrane localization in human cells
Abstract
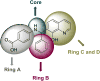
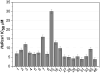
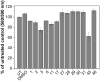
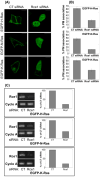
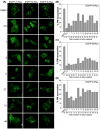
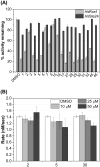
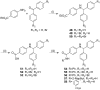
Ras converting enzyme 1 (Rce1) is an endoprotease that catalyzes processing of the C-terminus of Ras protein by removing -aaX from the CaaX motif. The activity of Rce1 is crucial for proper localization of Ras to the plasma membrane where it functions. Ras is responsible for transmitting signals related to cell proliferation, cell cycle progression, and apoptosis. The disregulation of these pathways due to constitutively active oncogenic Ras can ultimately lead to cancer. Ras, its effectors and regulators, and the enzymes that are involved in its maturation process are all targets for anti-cancer therapeutics. Key enzymes required for Ras maturation and localization are the farnesyltransferase (FTase), Rce1, and isoprenylcysteine carboxyl methyltransferase (ICMT). Among these proteins, the physiological role of Rce1 in regulating Ras and other CaaX proteins has not been fully explored. Small-molecule inhibitors of Rce1 could be useful as chemical biology tools to understand further the downstream impact of Rce1 on Ras function and serve as potential leads for cancer therapeutics. Structure-activity relationship (SAR) analysis of a previously reported Rce1 inhibitor, NSC1011, has been performed to generate a new library of Rce1 inhibitors. The new inhibitors caused a reduction in Rce1 in vitro activity, exhibited low cell toxicity, and induced mislocalization of EGFP-Ras from the plasma membrane in human colon carcinoma cells giving rise to a phenotype similar to that observed with siRNA knockdowns of Rce1 expression. Several of the new inhibitors were more effective at mislocalizing K-Ras compared to a potent farnesyltransferase inhibitor (FTI), which is significant because of the preponderance of K-Ras mutations in cancer.
Keywords: Protease inhibitors; Ras converting enzyme (Rce1); Ras mislocalization.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
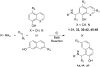
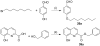
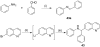
Similar articles
-
Rce1: mechanism and inhibition.Crit Rev Biochem Mol Biol. 2018 Apr;53(2):157-174. doi: 10.1080/10409238.2018.1431606. Epub 2018 Feb 9. Crit Rev Biochem Mol Biol. 2018. PMID: 29424242 Free PMC article. Review.
-
Absence of the CAAX endoprotease Rce1: effects on cell growth and transformation.Mol Cell Biol. 2002 Jan;22(1):171-81. doi: 10.1128/MCB.22.1.171-181.2002. Mol Cell Biol. 2002. PMID: 11739732 Free PMC article.
-
A novel RCE1 isoform is required for H-Ras plasma membrane localization and is regulated by USP17.Biochem J. 2014 Jan 15;457(2):289-300. doi: 10.1042/BJ20131213. Biochem J. 2014. PMID: 24134311
-
Endoproteolytic processing of RhoA by Rce1 is required for the cleavage of RhoA by Yersinia enterocolitica outer protein T.Infect Immun. 2006 Mar;74(3):1712-7. doi: 10.1128/IAI.74.3.1712-1717.2006. Infect Immun. 2006. PMID: 16495543 Free PMC article.
-
Inhibitors of chronically active ras: potential for treatment of human malignancies.Recent Pat Anticancer Drug Discov. 2008 Jan;3(1):31-47. doi: 10.2174/157489208783478702. Recent Pat Anticancer Drug Discov. 2008. PMID: 18289122 Review.
Cited by
-
A Humanized Yeast System for Evaluating the Protein Prenylation of a Wide Range of Human and Viral CaaX Sequences.bioRxiv [Preprint]. 2023 Sep 20:2023.09.19.558494. doi: 10.1101/2023.09.19.558494. bioRxiv. 2023. Update in: Dis Model Mech. 2024 May 1;17(5):dmm050516. doi: 10.1242/dmm.050516. PMID: 37786692 Free PMC article. Updated. Preprint.
-
Synthesis of Bioactive Aminomethylated 8-Hydroxyquinolines via the Modified Mannich Reaction.Int J Mol Sci. 2023 Apr 26;24(9):7915. doi: 10.3390/ijms24097915. Int J Mol Sci. 2023. PMID: 37175622 Free PMC article. Review.
-
RCE1 deficiency enhances invasion via the promotion of epithelial-mesenchymal transition and predicts poor prognosis in hepatocellular carcinoma.Am J Transl Res. 2020 Nov 15;12(11):7236-7248. eCollection 2020. Am J Transl Res. 2020. PMID: 33312363 Free PMC article.
-
Post-translational modification of KRAS: potential targets for cancer therapy.Acta Pharmacol Sin. 2021 Aug;42(8):1201-1211. doi: 10.1038/s41401-020-00542-y. Epub 2020 Oct 21. Acta Pharmacol Sin. 2021. PMID: 33087838 Free PMC article. Review.
-
A Novel KRAS Antibody Highlights a Regulation Mechanism of Post-Translational Modifications of KRAS during Tumorigenesis.Int J Mol Sci. 2020 Sep 2;21(17):6361. doi: 10.3390/ijms21176361. Int J Mol Sci. 2020. PMID: 32887255 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Miscellaneous

