Ubiquitin regulates TORC1 in yeast Saccharomyces cerevisiae
- PMID: 26700129
- PMCID: PMC6697181
- DOI: 10.1111/mmi.13319
Ubiquitin regulates TORC1 in yeast Saccharomyces cerevisiae
Abstract
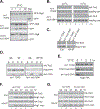
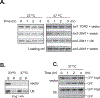
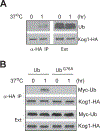
In the yeast Saccharomyces cerevisiae the TOR complex 1 (TORC1) controls many growth-related cellular processes and is essential for cell growth and proliferation. Macrolide antibiotic rapamycin, in complex with a cytosol protein named FKBP12, specifically inhibits TORC1, causing growth arrest. The FKBP12-rapamycin complex interferes with TORC1 function by binding to the FRB domain of the TOR proteins. In an attempt to understand the role of the FRB domain in TOR function, we identified a single point mutation (Tor2(W2041R) ) in the FRB domain of Tor2 that renders yeast cells rapamycin resistant and temperature sensitive. At the permissive temperature, the Tor2 mutant protein is partially defective for binding with Kog1 and TORC1 is impaired for membrane association. At the restrictive temperature, Kog1 but not the Tor2 mutant protein, is rapidly degraded. Overexpression of ubiquitin stabilizes Kog1 and suppresses the growth defect associated with the tor2 mutant at the nonpremissive temperature. We find that ubiquitin binds non-covalently to Kog1, prevents Kog1 from degradation and stabilizes TORC1. Our data reveal a unique role for ubiquitin in regulation of TORC1 and suggest that Kog1 requires association with the Tor proteins for stabilization.
© 2016 John Wiley & Sons Ltd.
Figures



Similar articles
-
Regulation of TORC1 by ubiquitin through non-covalent binding.Curr Genet. 2016 Aug;62(3):553-5. doi: 10.1007/s00294-016-0581-7. Epub 2016 Feb 24. Curr Genet. 2016. PMID: 26910532 Free PMC article. Review.
-
Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control.Mol Cell. 2002 Sep;10(3):457-68. doi: 10.1016/s1097-2765(02)00636-6. Mol Cell. 2002. PMID: 12408816
-
Structure of TOR and its complex with KOG1.Mol Cell. 2007 Aug 3;27(3):509-16. doi: 10.1016/j.molcel.2007.05.040. Mol Cell. 2007. PMID: 17679098
-
LAS24/KOG1, a component of the TOR complex 1 (TORC1), is needed for resistance to local anesthetic tetracaine and normal distribution of actin cytoskeleton in yeast.Genes Genet Syst. 2005 Oct;80(5):325-43. doi: 10.1266/ggs.80.325. Genes Genet Syst. 2005. PMID: 16394584
-
Evolutionary Conservation of the Components in the TOR Signaling Pathways.Biomolecules. 2017 Nov 1;7(4):77. doi: 10.3390/biom7040077. Biomolecules. 2017. PMID: 29104218 Free PMC article. Review.
Cited by
-
TORC1 Signaling in Fungi: From Yeasts to Filamentous Fungi.Microorganisms. 2023 Jan 15;11(1):218. doi: 10.3390/microorganisms11010218. Microorganisms. 2023. PMID: 36677510 Free PMC article. Review.
-
Investigating the Antifungal Mechanism of Action of Polygodial by Phenotypic Screening in Saccharomyces cerevisiae.Int J Mol Sci. 2021 May 28;22(11):5756. doi: 10.3390/ijms22115756. Int J Mol Sci. 2021. PMID: 34071169 Free PMC article.
-
Regulation of TORC1 by ubiquitin through non-covalent binding.Curr Genet. 2016 Aug;62(3):553-5. doi: 10.1007/s00294-016-0581-7. Epub 2016 Feb 24. Curr Genet. 2016. PMID: 26910532 Free PMC article. Review.
-
TOR-autophagy branch signaling via Imp1 dictates plant-microbe biotrophic interface longevity.PLoS Genet. 2018 Nov 21;14(11):e1007814. doi: 10.1371/journal.pgen.1007814. eCollection 2018 Nov. PLoS Genet. 2018. PMID: 30462633 Free PMC article.
-
The nutrient transceptor/PKA pathway functions independently of TOR and responds to leucine and Gcn2 in a TOR-independent manner.FEMS Yeast Res. 2017 Aug 1;17(5):fox048. doi: 10.1093/femsyr/fox048. FEMS Yeast Res. 2017. PMID: 28810702 Free PMC article.
References
-
- Adami A, Garcia-Alvarez B, Arias-Palomo E, Barford D & Llorca O, (2007) Structure of TOR and its complex with KOG1. Molecular cell 27: 509–516. - PubMed
-
- Choi J, Chen J, Schreiber SL & Clardy J, (1996) Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273: 239–242. - PubMed
-
- Ecker DJ, Khan MI, Marsh J, Butt TR & Crooke ST, (1987) Chemical synthesis and expression of a cassette adapted ubiquitin gene. J Biol Chem 262: 3524–3527. - PubMed
-
- Finley D, Ozkaynak E & Varshavsky A, (1987) The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48: 1035–1046. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

