VHICA, a New Method to Discriminate between Vertical and Horizontal Transposon Transfer: Application to the Mariner Family within Drosophila
- PMID: 26685176
- PMCID: PMC4776708
- DOI: 10.1093/molbev/msv341
VHICA, a New Method to Discriminate between Vertical and Horizontal Transposon Transfer: Application to the Mariner Family within Drosophila
Abstract
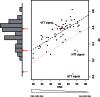
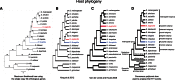
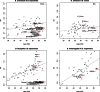
Transposable elements (TEs) are genomic repeated sequences that display complex evolutionary patterns. They are usually inherited vertically, but can occasionally be transmitted between sexually independent species, through so-called horizontal transposon transfers (HTTs). Recurrent HTTs are supposed to be essential in life cycle of TEs, which are otherwise destined for eventual decay. HTTs also impact the host genome evolution. However, the extent of HTTs in eukaryotes is largely unknown, due to the lack of efficient, statistically supported methods that can be applied to multiple species sequence data sets. Here, we developed a new automated method available as a R package "vhica" that discriminates whether a given TE family was vertically or horizontally transferred, and potentially infers donor and receptor species. The method is well suited for TE sequences extracted from complete genomes, and applicable to multiple TEs and species at the same time. We first validated our method using Drosophila TE families with well-known evolutionary histories, displaying both HTTs and vertical transmission. We then tested 26 different lineages of mariner elements recently characterized in 20 Drosophila genomes, and found HTTs in 24 of them. Furthermore, several independent HTT events could often be detected within the same mariner lineage. The VHICA (Vertical and Horizontal Inheritance Consistence Analysis) method thus appears as a valuable tool to analyze the evolutionary history of TEs across a large range of species.
Keywords: Drosophila; codon usage; horizontal transfer; mariner element; synonymous substitutions; transposable elements; vertical transmission.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



Similar articles
-
Helena and BS: Two Travellers between the Genera Drosophila and Zaprionus.Genome Biol Evol. 2018 Oct 1;10(10):2671-2685. doi: 10.1093/gbe/evy184. Genome Biol Evol. 2018. PMID: 30165545 Free PMC article.
-
Genomic landscape and evolutionary dynamics of mariner transposable elements within the Drosophila genus.BMC Genomics. 2014 Aug 27;15(1):727. doi: 10.1186/1471-2164-15-727. BMC Genomics. 2014. PMID: 25163909 Free PMC article.
-
Multiple events of horizontal transfer of the Minos transposable element between Drosophila species.Mol Phylogenet Evol. 2005 Jun;35(3):583-94. doi: 10.1016/j.ympev.2004.11.026. Epub 2005 Feb 1. Mol Phylogenet Evol. 2005. PMID: 15878127
-
Horizontal transfers of transposable elements in eukaryotes: The flying genes.C R Biol. 2016 Jul-Aug;339(7-8):296-9. doi: 10.1016/j.crvi.2016.04.013. Epub 2016 May 24. C R Biol. 2016. PMID: 27234293 Review.
-
[Transposable element mariner].Yi Chuan. 2004 Sep;26(5):756-62. Yi Chuan. 2004. PMID: 15640098 Review. Chinese.
Cited by
-
Identification of Bari Transposons in 23 Sequenced Drosophila Genomes Reveals Novel Structural Variants, MITEs and Horizontal Transfer.PLoS One. 2016 May 23;11(5):e0156014. doi: 10.1371/journal.pone.0156014. eCollection 2016. PLoS One. 2016. PMID: 27213270 Free PMC article.
-
An Analysis of IS630/Tc1/mariner Transposons in the Genome of a Pacific Oyster, Crassostrea gigas.J Mol Evol. 2018 Oct;86(8):566-580. doi: 10.1007/s00239-018-9868-2. Epub 2018 Oct 3. J Mol Evol. 2018. PMID: 30283979
-
Transposable Elements Co-Option in Genome Evolution and Gene Regulation.Int J Mol Sci. 2023 Jan 30;24(3):2610. doi: 10.3390/ijms24032610. Int J Mol Sci. 2023. PMID: 36768929 Free PMC article. Review.
-
Helena and BS: Two Travellers between the Genera Drosophila and Zaprionus.Genome Biol Evol. 2018 Oct 1;10(10):2671-2685. doi: 10.1093/gbe/evy184. Genome Biol Evol. 2018. PMID: 30165545 Free PMC article.
-
Horizontal acquisition of transposable elements and viral sequences: patterns and consequences.Curr Opin Genet Dev. 2018 Apr;49:15-24. doi: 10.1016/j.gde.2018.02.007. Epub 2018 Mar 2. Curr Opin Genet Dev. 2018. PMID: 29505963 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

