Role of the Interdomain Linker in RNA-Activated Protein Kinase Activation
- PMID: 26678943
- PMCID: PMC4718896
- DOI: 10.1021/acs.biochem.5b01171
Role of the Interdomain Linker in RNA-Activated Protein Kinase Activation
Abstract
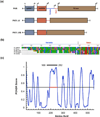
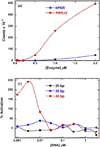
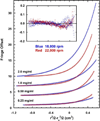
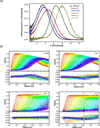
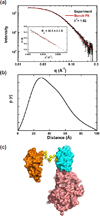
RNA-activated protein kinase (PKR) is a key component of the interferon-induced antiviral pathway in higher eukaryotes. Upon recognition of viral dsRNA, PKR is activated via dimerization and autophosphorylation. PKR contains two N-terminal dsRNA binding domains (dsRBD) and a C-terminal kinase domain. The dsRBDs and the kinase are separated by a long, unstructured ∼80-amino acid linker in the human enzyme. The length of the N-terminal portion of the linker varies among PKR sequences, and it is completely absent in one ortholog. Here, we characterize the effects of deleting the variable region from the human enzyme to produce PKRΔV. The linker deletion results in quantitative but not qualitative changes in catalytic activity, RNA binding, and conformation. PKRΔV is somewhat more active and exhibits more cooperative RNA binding. As we previously observed for the full-length enzyme, PKRΔV is flexible in solution and adopts a range of compact and extended conformations. The conformational ensemble is biased toward compact states that might be related to weak interactions between the dsRBD and kinase domains. PKR retains RNA-induced autophosphorylation upon complete removal of the linker, indicating that the C-terminal, basic region is also not required for activity.
Figures

Similar articles
-
Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs.J Mol Biol. 2006 May 19;358(5):1270-85. doi: 10.1016/j.jmb.2006.03.003. Epub 2006 Mar 20. J Mol Biol. 2006. PMID: 16580685
-
Mechanism of interaction of the double-stranded RNA (dsRNA) binding domain of protein kinase R with short dsRNA sequences.Biochemistry. 2007 Jan 9;46(1):55-65. doi: 10.1021/bi061531o. Biochemistry. 2007. PMID: 17198375
-
Molecular framework for the activation of RNA-dependent protein kinase.J Biol Chem. 2007 Apr 13;282(15):11474-86. doi: 10.1074/jbc.M700301200. Epub 2007 Feb 6. J Biol Chem. 2007. PMID: 17284445
-
Activation of PKR: an open and shut case?Trends Biochem Sci. 2007 Feb;32(2):57-62. doi: 10.1016/j.tibs.2006.12.003. Epub 2006 Dec 29. Trends Biochem Sci. 2007. PMID: 17196820 Free PMC article. Review.
-
PKR and eIF2alpha: integration of kinase dimerization, activation, and substrate docking.Cell. 2005 Sep 23;122(6):823-5. doi: 10.1016/j.cell.2005.09.007. Cell. 2005. PMID: 16179248 Review.
Cited by
-
Examination of yield, bacteriolytic activity and cold storage of linker deletion mutants based on endolysin S6_ORF93 derived from Staphylococcus giant bacteriophage S6.PLoS One. 2024 Oct 23;19(10):e0310962. doi: 10.1371/journal.pone.0310962. eCollection 2024. PLoS One. 2024. PMID: 39441843 Free PMC article.
-
The search for a PKR code-differential regulation of protein kinase R activity by diverse RNA and protein regulators.RNA. 2019 May;25(5):539-556. doi: 10.1261/rna.070169.118. Epub 2019 Feb 15. RNA. 2019. PMID: 30770398 Free PMC article. Review.
-
Auto-phosphorylation Represses Protein Kinase R Activity.Sci Rep. 2017 Mar 10;7:44340. doi: 10.1038/srep44340. Sci Rep. 2017. PMID: 28281686 Free PMC article.
References
-
- Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. - PubMed
-
- Unterholzner L, Bowie AG. The interplay between viruses and innate immune signaling: recent insights and therapeutic opportunities. Biochem. Pharmacol. 2008;75:589–602. - PubMed
-
- Pindel A, Sadler A. The role of protein kinase R in the interferon response. J Interferon Cytokine Res. 2011;31:59–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

