Treatment of Triple-Negative Breast Cancer with TORC1/2 Inhibitors Sustains a Drug-Resistant and Notch-Dependent Cancer Stem Cell Population
- PMID: 26676751
- PMCID: PMC4715956
- DOI: 10.1158/0008-5472.CAN-15-1640-T
Treatment of Triple-Negative Breast Cancer with TORC1/2 Inhibitors Sustains a Drug-Resistant and Notch-Dependent Cancer Stem Cell Population
Erratum in
-
Correction: Treatment of Triple-Negative Breast Cancer with TORC1/2 Inhibitors Sustains a Drug-Resistant and Notch-Dependent Cancer Stem Cell Population.Cancer Res. 2019 Feb 15;79(4):875. doi: 10.1158/0008-5472.CAN-18-4087. Cancer Res. 2019. PMID: 30770370 No abstract available.
Abstract
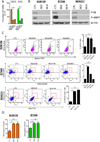
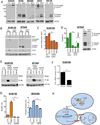
Approximately 30% of triple-negative breast cancers (TNBC) harbor molecular alterations in PI3K/mTOR signaling, but therapeutic inhibition of this pathway has not been effective. We hypothesized that intrinsic resistance to TORC1/2 inhibition is driven by cancer stem cell (CSC)-like populations that could be targeted to enhance the antitumor action of these drugs. Therefore, we investigated the molecular mechanisms by which PI3K/mTOR inhibitors affect the stem-like properties of TNBC cells. Treatment of established TNBC cell lines with a PI3K/mTOR inhibitor or a TORC1/2 inhibitor increased the expression of CSC markers and mammosphere formation. A CSC-specific PCR array revealed that inhibition of TORC1/2 increased FGF1 and Notch1 expression. Notch1 activity was also induced in TNBC cells treated with TORC1/2 inhibitors and associated with increased mitochondrial metabolism and FGFR1 signaling. Notably, genetic and pharmacologic blockade of Notch1 abrogated the increase in CSC markers, mammosphere formation, and in vivo tumor-initiating capacity induced by TORC1/2 inhibition. These results suggest that targeting the FGFR-mitochondrial metabolism-Notch1 axis prevents resistance to TORC1/2 inhibitors by eradicating drug-resistant CSCs in TNBC, and may thus represent an attractive therapeutic strategy to improve drug responsiveness and efficacy.
©2015 American Association for Cancer Research.
Conflict of interest statement
The authors in this manuscript have no conflicts of interest to declare.
Figures




Similar articles
-
Patient-derived xenografts of triple-negative breast cancer reproduce molecular features of patient tumors and respond to mTOR inhibition.Breast Cancer Res. 2014 Apr 7;16(2):R36. doi: 10.1186/bcr3640. Breast Cancer Res. 2014. PMID: 24708766 Free PMC article.
-
BCL6 and the Notch pathway: a signaling axis leading to a novel druggable biotarget in triple negative breast cancer.Cell Oncol (Dordr). 2022 Apr;45(2):257-274. doi: 10.1007/s13402-022-00663-y. Epub 2022 Mar 31. Cell Oncol (Dordr). 2022. PMID: 35357654
-
c-Jun N-terminal kinase promotes stem cell phenotype in triple-negative breast cancer through upregulation of Notch1 via activation of c-Jun.Oncogene. 2017 May 4;36(18):2599-2608. doi: 10.1038/onc.2016.417. Epub 2016 Dec 12. Oncogene. 2017. PMID: 27941886 Free PMC article.
-
PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors.Breast Cancer Res. 2014 Aug 8;16(4):406. doi: 10.1186/s13058-014-0406-x. Breast Cancer Res. 2014. PMID: 25103565 Free PMC article.
-
Targeting the PI3K/AKT/mTOR pathway in triple-negative breast cancer: a review.Breast Cancer Res Treat. 2018 Jun;169(3):397-406. doi: 10.1007/s10549-018-4697-y. Epub 2018 Feb 7. Breast Cancer Res Treat. 2018. PMID: 29417298 Review.
Cited by
-
Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies.Front Immunol. 2020 Aug 7;11:1280. doi: 10.3389/fimmu.2020.01280. eCollection 2020. Front Immunol. 2020. PMID: 32849491 Free PMC article. Review.
-
Triple negative breast cancer: shedding light onto the role of pi3k/akt/mtor pathway.Oncotarget. 2016 Sep 13;7(37):60712-60722. doi: 10.18632/oncotarget.10858. Oncotarget. 2016. PMID: 27474173 Free PMC article. Review.
-
Moving Breast Cancer Therapy up a Notch.Front Oncol. 2018 Nov 20;8:518. doi: 10.3389/fonc.2018.00518. eCollection 2018. Front Oncol. 2018. PMID: 30515368 Free PMC article. Review.
-
The Efficacy of CB-103, a First-in-Class Transcriptional Notch Inhibitor, in Preclinical Models of Breast Cancer.Cancers (Basel). 2023 Aug 3;15(15):3957. doi: 10.3390/cancers15153957. Cancers (Basel). 2023. PMID: 37568775 Free PMC article.
-
The Missing Lnc: The Potential of Targeting Triple-Negative Breast Cancer and Cancer Stem Cells by Inhibiting Long Non-Coding RNAs.Cells. 2020 Mar 20;9(3):763. doi: 10.3390/cells9030763. Cells. 2020. PMID: 32244924 Free PMC article. Review.
References
-
- Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, Viens P, et al. How basal are triple-negative breast cancers? International Journal of Cancer. 2008;123:236–240. - PubMed
-
- Liedtke C, Mazouni C, Hess K, Andre F, Tordai A, Mejia J, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. - PubMed
-
- Shah S, Morin R, Khattra J, Prentice L, Pugh T, Burleigh A, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

