Regulation of gene transcription by Polycomb proteins
- PMID: 26665172
- PMCID: PMC4672759
- DOI: 10.1126/sciadv.1500737
Regulation of gene transcription by Polycomb proteins
Abstract
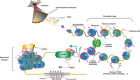
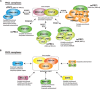
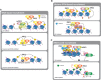
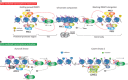
The Polycomb group (PcG) of proteins defines a subset of factors that physically associate and function to maintain the positional identity of cells from the embryo to adult stages. PcG has long been considered a paradigmatic model for epigenetic maintenance of gene transcription programs. Despite intensive research efforts to unveil the molecular mechanisms of action of PcG proteins, several fundamental questions remain unresolved: How many different PcG complexes exist in mammalian cells? How are PcG complexes targeted to specific loci? How does PcG regulate transcription? In this review, we discuss the diversity of PcG complexes in mammalian cells, examine newly identified modes of recruitment to chromatin, and highlight the latest insights into the molecular mechanisms underlying the function of PcGs in transcription regulation and three-dimensional chromatin conformation.
Keywords: Chromatin; Epigenetics; Gene Regulation.
Figures
Similar articles
-
One, Two, Three: Polycomb Proteins Hit All Dimensions of Gene Regulation.Genes (Basel). 2015 Jul 10;6(3):520-42. doi: 10.3390/genes6030520. Genes (Basel). 2015. PMID: 26184319 Free PMC article. Review.
-
Variations on a theme: Polycomb group proteins in plants.J Exp Bot. 2014 Jun;65(10):2769-84. doi: 10.1093/jxb/ert410. Epub 2013 Dec 13. J Exp Bot. 2014. PMID: 24336446 Review.
-
Distinct roles of Polycomb group gene products in transcriptionally repressed and active domains of Hoxb8.Development. 2006 Jun;133(12):2371-81. doi: 10.1242/dev.02405. Epub 2006 May 10. Development. 2006. PMID: 16687444
-
Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1.Mol Cell Biol. 2005 Dec;25(24):11047-58. doi: 10.1128/MCB.25.24.11047-11058.2005. Mol Cell Biol. 2005. PMID: 16314526 Free PMC article.
-
The age of multiplexity: recruitment and interactions of Polycomb complexes in plants.Curr Opin Plant Biol. 2016 Feb;29:169-78. doi: 10.1016/j.pbi.2015.11.010. Epub 2016 Jan 28. Curr Opin Plant Biol. 2016. PMID: 26826786 Review.
Cited by
-
Ubiquitin-dependent regulation of transcription in development and disease.EMBO Rep. 2021 Apr 7;22(4):e51078. doi: 10.15252/embr.202051078. Epub 2021 Mar 28. EMBO Rep. 2021. PMID: 33779035 Free PMC article. Review.
-
The Systematic Analyses of RING Finger Gene Signature for Predicting the Prognosis of Patients with Hepatocellular Carcinoma.J Oncol. 2022 Sep 26;2022:2466006. doi: 10.1155/2022/2466006. eCollection 2022. J Oncol. 2022. PMID: 36199791 Free PMC article.
-
Chromatin dynamics associated with seed desiccation tolerance/sensitivity at early germination in Medicago truncatula.Front Plant Sci. 2022 Nov 24;13:1059493. doi: 10.3389/fpls.2022.1059493. eCollection 2022. Front Plant Sci. 2022. PMID: 36507374 Free PMC article.
-
Polycomb protein RYBP activates transcription factor Plagl1 during in vitro cardiac differentiation of mouse embryonic stem cells.Open Biol. 2023 Feb;13(2):220305. doi: 10.1098/rsob.220305. Epub 2023 Feb 8. Open Biol. 2023. PMID: 36751888 Free PMC article.
-
Histone variants and modifications during abiotic stress response.Front Plant Sci. 2022 Dec 15;13:984702. doi: 10.3389/fpls.2022.984702. eCollection 2022. Front Plant Sci. 2022. PMID: 36589114 Free PMC article. Review.
References
-
- W. Flemming, Zellsubstanz, Kern und Zelltheilung (F. C. W. Vogel, Leipzig, 1882).
-
- Kouzarides T., Chromatin modifications and their function. Cell 128, 693–705 (2007). - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

