Towards a predictive model of chromatin 3D organization
- PMID: 26658098
- PMCID: PMC4892986
- DOI: 10.1016/j.semcdb.2015.11.013
Towards a predictive model of chromatin 3D organization
Abstract
Architectural proteins mediate interactions between distant regions in the genome to bring together different regulatory elements while establishing a specific three-dimensional organization of the genetic material. Depletion of specific architectural proteins leads to miss regulation of gene expression and alterations in nuclear organization. The specificity of interactions mediated by architectural proteins depends on the nature, number, and orientation of their binding site at individual genomic locations. Knowledge of the mechanisms and rules governing interactions among architectural proteins may provide a code to predict the 3D organization of the genome.
Keywords: Architectural proteins; CTCF; Nucleus; Transcription.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures

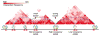
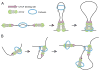
Similar articles
-
Architectural proteins, transcription, and the three-dimensional organization of the genome.FEBS Lett. 2015 Oct 7;589(20 Pt A):2923-30. doi: 10.1016/j.febslet.2015.05.025. Epub 2015 May 22. FEBS Lett. 2015. PMID: 26008126 Free PMC article. Review.
-
DNA Supercoiling, Topoisomerases, and Cohesin: Partners in Regulating Chromatin Architecture?Int J Mol Sci. 2018 Mar 16;19(3):884. doi: 10.3390/ijms19030884. Int J Mol Sci. 2018. PMID: 29547555 Free PMC article. Review.
-
Architectural proteins: regulators of 3D genome organization in cell fate.Trends Cell Biol. 2014 Nov;24(11):703-11. doi: 10.1016/j.tcb.2014.08.003. Epub 2014 Sep 10. Trends Cell Biol. 2014. PMID: 25218583 Free PMC article. Review.
-
CTCF Binding Polarity Determines Chromatin Looping.Mol Cell. 2015 Nov 19;60(4):676-84. doi: 10.1016/j.molcel.2015.09.023. Epub 2015 Oct 29. Mol Cell. 2015. PMID: 26527277
-
CTCF as a multifunctional protein in genome regulation and gene expression.Exp Mol Med. 2015 Jun 5;47(6):e166. doi: 10.1038/emm.2015.33. Exp Mol Med. 2015. PMID: 26045254 Free PMC article. Review.
Cited by
-
Computational approaches for inferring 3D conformations of chromatin from chromosome conformation capture data.Methods. 2020 Oct 1;181-182:24-34. doi: 10.1016/j.ymeth.2019.08.008. Epub 2019 Aug 27. Methods. 2020. PMID: 31470090 Free PMC article. Review.
-
Evolutionary conserved NSL complex/BRD4 axis controls transcription activation via histone acetylation.Nat Commun. 2020 May 7;11(1):2243. doi: 10.1038/s41467-020-16103-0. Nat Commun. 2020. PMID: 32382029 Free PMC article.
-
Machine and Deep Learning Methods for Predicting 3D Genome Organization.Methods Mol Biol. 2025;2856:357-400. doi: 10.1007/978-1-0716-4136-1_22. Methods Mol Biol. 2025. PMID: 39283464 Review.
-
Integrating epigenomic data and 3D genomic structure with a new measure of chromatin assortativity.Genome Biol. 2016 Jul 8;17(1):152. doi: 10.1186/s13059-016-1003-3. Genome Biol. 2016. PMID: 27391817 Free PMC article.
-
5C analysis of the Epidermal Differentiation Complex locus reveals distinct chromatin interaction networks between gene-rich and gene-poor TADs in skin epithelial cells.PLoS Genet. 2017 Sep 1;13(9):e1006966. doi: 10.1371/journal.pgen.1006966. eCollection 2017 Sep. PLoS Genet. 2017. PMID: 28863138 Free PMC article.
References
-
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

