Preassembled Single-Stranded RNA-Argonaute Complexes: A Novel Method to Silence Genes in Cryptosporidium
- PMID: 26656125
- PMCID: PMC4799669
- DOI: 10.1093/infdis/jiv588
Preassembled Single-Stranded RNA-Argonaute Complexes: A Novel Method to Silence Genes in Cryptosporidium
Abstract
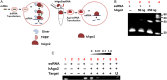
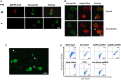
Cryptosporidiosis is a common cause of diarrhea morbidity and mortality worldwide. Research progress on this infection has been slowed by lack of methods to genetically manipulate Cryptosporidium parasites. Small interfering RNA (siRNA) is widely used to study gene function, but Cryptosporidium species lack the enzymes necessary to process siRNA. By preassembling complexes with the human enzyme Argonaute 2 (hAgo2) and Cryptosporidium single-stranded RNA (ssRNA), we induced specific slicing in Cryptosporidium RNA targets. We demonstrated the reduction in expression of target genes at the mRNA and protein levels by transfecting live parasites with ssRNA-hAgo2 complexes. Furthermore we used this method to confirm the role of selected molecules during host cell invasion. This novel method provides a novel means of silencing Cryptosporidium genes to study their role in host-parasite interactions and as potential targets for chemotherapy.
Keywords: Argonaute; Cryptosporidium; Cryptosporidium parvum; cryptosporidiosis; siRNA; silencing.
© The Author 2015. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail journals.permissions@oup.com.
Figures


Similar articles
-
RNA-Based Therapy for Cryptosporidium parvum Infection: Proof-of-Concept Studies.Infect Immun. 2022 Jul 21;90(7):e0019622. doi: 10.1128/iai.00196-22. Epub 2022 Jun 1. Infect Immun. 2022. PMID: 35647663 Free PMC article.
-
A Novel Method to Silence Genes in Cryptosporidium.Methods Mol Biol. 2020;2052:193-203. doi: 10.1007/978-1-4939-9748-0_11. Methods Mol Biol. 2020. PMID: 31452163 Free PMC article.
-
Recent advances in genetic manipulation of Cryptosporidium.Curr Opin Microbiol. 2020 Dec;58:146-152. doi: 10.1016/j.mib.2020.09.010. Epub 2020 Nov 5. Curr Opin Microbiol. 2020. PMID: 33161368 Free PMC article. Review.
-
Transcriptome analysis of pig intestinal cell monolayers infected with Cryptosporidium parvum asexual stages.Parasit Vectors. 2018 Mar 12;11(1):176. doi: 10.1186/s13071-018-2754-3. Parasit Vectors. 2018. PMID: 29530089 Free PMC article.
-
Cryptosporidium: host and parasite transcriptome in infection.Curr Opin Microbiol. 2020 Dec;58:138-145. doi: 10.1016/j.mib.2020.09.012. Epub 2020 Nov 4. Curr Opin Microbiol. 2020. PMID: 33160225 Review.
Cited by
-
Exploring the impact of digestive physicochemical parameters of adults and infants on the pathophysiology of Cryptosporidium parvum using the dynamic TIM-1 gastrointestinal model.Gut Pathog. 2024 Oct 1;16(1):55. doi: 10.1186/s13099-024-00648-2. Gut Pathog. 2024. PMID: 39354600 Free PMC article.
-
A tiny loop in the Argonaute PIWI domain tunes small RNA seed strength.EMBO Rep. 2023 Jun 5;24(6):e55806. doi: 10.15252/embr.202255806. Epub 2023 Apr 21. EMBO Rep. 2023. PMID: 37082939 Free PMC article.
-
RNA-Based Therapy for Cryptosporidium parvum Infection: Proof-of-Concept Studies.Infect Immun. 2022 Jul 21;90(7):e0019622. doi: 10.1128/iai.00196-22. Epub 2022 Jun 1. Infect Immun. 2022. PMID: 35647663 Free PMC article.
-
A Novel Calcium-Dependent Kinase Inhibitor, Bumped Kinase Inhibitor 1517, Cures Cryptosporidiosis in Immunosuppressed Mice.J Infect Dis. 2016 Dec 15;214(12):1850-1855. doi: 10.1093/infdis/jiw481. Epub 2016 Oct 12. J Infect Dis. 2016. PMID: 27738055 Free PMC article.
-
Recent Breakthroughs and Ongoing Limitations in Cryptosporidium Research.F1000Res. 2018 Sep 3;7:F1000 Faculty Rev-1380. doi: 10.12688/f1000research.15333.1. eCollection 2018. F1000Res. 2018. PMID: 30228873 Free PMC article. Review.
References
-
- Kotloff KL, Nataro JP, Blackwelder WC et al. . Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013; 382:209–22. - PubMed
-
- Xu P, Widmer G, Wang Y et al. . The genome of Cryptosporidium hominis. Nature 2004; 431:1107–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

