Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis
- PMID: 26655835
- PMCID: PMC4844191
- DOI: 10.1038/ncb3289
Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis
Abstract
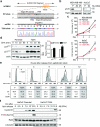
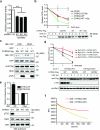
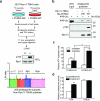
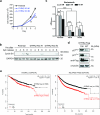
Despite the fundamental importance of proteasomal degradation in cells, little is known about whether and how the 26S proteasome itself is regulated in coordination with various physiological processes. Here we show that the proteasome is dynamically phosphorylated during the cell cycle at Thr 25 of the 19S subunit Rpt3. CRISPR/Cas9-mediated genome editing, RNA interference and biochemical studies demonstrate that blocking Rpt3-Thr25 phosphorylation markedly impairs proteasome activity and impedes cell proliferation. Through a kinome-wide screen, we have identified dual-specificity tyrosine-regulated kinase 2 (DYRK2) as the primary kinase that phosphorylates Rpt3-Thr25, leading to enhanced substrate translocation and degradation. Importantly, loss of the single phosphorylation of Rpt3-Thr25 or knockout of DYRK2 significantly inhibits tumour formation by proteasome-addicted human breast cancer cells in mice. These findings define an important mechanism for proteasome regulation and demonstrate the biological significance of proteasome phosphorylation in regulating cell proliferation and tumorigenesis.
Figures



Comment in
-
Ramping up degradation for proliferation.Nat Cell Biol. 2016 Feb;18(2):141-2. doi: 10.1038/ncb3306. Nat Cell Biol. 2016. PMID: 26820437
-
The 26S proteasome: A cell cycle regulator regulated by cell cycle.Cell Cycle. 2016;15(7):875-6. doi: 10.1080/15384101.2016.1151728. Epub 2016 Mar 3. Cell Cycle. 2016. PMID: 26940127 Free PMC article. No abstract available.
Similar articles
-
Ramping up degradation for proliferation.Nat Cell Biol. 2016 Feb;18(2):141-2. doi: 10.1038/ncb3306. Nat Cell Biol. 2016. PMID: 26820437
-
Ancient drug curcumin impedes 26S proteasome activity by direct inhibition of dual-specificity tyrosine-regulated kinase 2.Proc Natl Acad Sci U S A. 2018 Aug 7;115(32):8155-8160. doi: 10.1073/pnas.1806797115. Epub 2018 Jul 9. Proc Natl Acad Sci U S A. 2018. PMID: 29987021 Free PMC article.
-
Diminished DYRK2 sensitizes hormone receptor-positive breast cancer to everolimus by the escape from degrading mTOR.Cancer Lett. 2017 Jan 1;384:27-38. doi: 10.1016/j.canlet.2016.10.015. Epub 2016 Oct 13. Cancer Lett. 2017. PMID: 27746162
-
Regulating protein breakdown through proteasome phosphorylation.Biochem J. 2017 Sep 24;474(19):3355-3371. doi: 10.1042/BCJ20160809. Biochem J. 2017. PMID: 28947610 Free PMC article. Review.
-
Tribbles-Related Protein Family Members as Regulators or Substrates of the Ubiquitin-Proteasome System in Cancer Development.Curr Cancer Drug Targets. 2016;16(2):147-56. doi: 10.2174/1568009616666151112122645. Curr Cancer Drug Targets. 2016. PMID: 26560117 Review.
Cited by
-
Proteasome condensate formation is driven by multivalent interactions with shuttle factors and ubiquitin chains.Proc Natl Acad Sci U S A. 2024 Mar 5;121(10):e2310756121. doi: 10.1073/pnas.2310756121. Epub 2024 Feb 26. Proc Natl Acad Sci U S A. 2024. PMID: 38408252 Free PMC article.
-
Genome-wide CRISPR Screen to Identify Genes that Suppress Transformation in the Presence of Endogenous KrasG12D.Sci Rep. 2019 Nov 20;9(1):17220. doi: 10.1038/s41598-019-53572-w. Sci Rep. 2019. PMID: 31748650 Free PMC article.
-
Functional screening of amplification outlier oncogenes in organoid models of early tumorigenesis.Cell Rep. 2023 Nov 28;42(11):113355. doi: 10.1016/j.celrep.2023.113355. Epub 2023 Nov 1. Cell Rep. 2023. PMID: 37922313 Free PMC article.
-
Thermal proteome profiling of breast cancer cells reveals proteasomal activation by CDK4/6 inhibitor palbociclib.EMBO J. 2018 May 15;37(10):e98359. doi: 10.15252/embj.201798359. Epub 2018 Apr 18. EMBO J. 2018. PMID: 29669860 Free PMC article.
-
RNF212B E3 ligase is essential for crossover designation and maturation during male and female meiosis in the mouse.Proc Natl Acad Sci U S A. 2024 Jun 18;121(25):e2320995121. doi: 10.1073/pnas.2320995121. Epub 2024 Jun 12. Proc Natl Acad Sci U S A. 2024. PMID: 38865271 Free PMC article.
References
-
- Coux O, Tanaka K, Goldberg AL. Structure and Functions of the 20S and 26S Proteasomes. Annual Review of Biochemistry. 1996;65:801–847. - PubMed
-
- Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nature reviews. Neuroscience. 2008;9:826–838. - PubMed
-
- Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. - PubMed
-
- Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res. 2008;14:1649–1657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

