Meta- and Orthogonal Integration of Influenza "OMICs" Data Defines a Role for UBR4 in Virus Budding
- PMID: 26651948
- PMCID: PMC4829074
- DOI: 10.1016/j.chom.2015.11.002
Meta- and Orthogonal Integration of Influenza "OMICs" Data Defines a Role for UBR4 in Virus Budding
Abstract
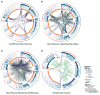
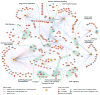
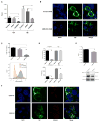
Several systems-level datasets designed to dissect host-pathogen interactions during influenza A infection have been reported. However, apparent discordance among these data has hampered their full utility toward advancing mechanistic and therapeutic knowledge. To collectively reconcile these datasets, we performed a meta-analysis of data from eight published RNAi screens and integrated these data with three protein interaction datasets, including one generated within the context of this study. Further integration of these data with global virus-host interaction analyses revealed a functionally validated biochemical landscape of the influenza-host interface, which can be queried through a simplified and customizable web portal (http://www.metascape.org/IAV). Follow-up studies revealed that the putative ubiquitin ligase UBR4 associates with the viral M2 protein and promotes apical transport of viral proteins. Taken together, the integrative analysis of influenza OMICs datasets illuminates a viral-host network of high-confidence human proteins that are essential for influenza A virus replication.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Gambling with Flu: "All in" to Maximize Reward.Cell Host Microbe. 2015 Dec 9;18(6):643-5. doi: 10.1016/j.chom.2015.11.010. Cell Host Microbe. 2015. PMID: 26651939
Similar articles
-
Host Cellular Protein TRAPPC6AΔ Interacts with Influenza A Virus M2 Protein and Regulates Viral Propagation by Modulating M2 Trafficking.J Virol. 2016 Dec 16;91(1):e01757-16. doi: 10.1128/JVI.01757-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795429 Free PMC article.
-
Nucleolar Relocalization of RBM14 by Influenza A Virus NS1 Protein.mSphere. 2018 Nov 14;3(6):e00549-18. doi: 10.1128/mSphereDirect.00549-18. mSphere. 2018. PMID: 30429226 Free PMC article.
-
A Defect in Influenza A Virus Particle Assembly Specific to Primary Human Macrophages.mBio. 2018 Oct 23;9(5):e01916-18. doi: 10.1128/mBio.01916-18. mBio. 2018. PMID: 30352935 Free PMC article.
-
Influenza A Virus M2 Protein: Roles from Ingress to Egress.Int J Mol Sci. 2017 Dec 7;18(12):2649. doi: 10.3390/ijms18122649. Int J Mol Sci. 2017. PMID: 29215568 Free PMC article. Review.
-
Microtubules in Influenza Virus Entry and Egress.Viruses. 2020 Jan 17;12(1):117. doi: 10.3390/v12010117. Viruses. 2020. PMID: 31963544 Free PMC article. Review.
Cited by
-
Binding of nuclear factor κB to noncanonical consensus sites reveals its multimodal role during the early inflammatory response.Genome Res. 2016 Nov;26(11):1478-1489. doi: 10.1101/gr.210005.116. Epub 2016 Sep 15. Genome Res. 2016. PMID: 27633323 Free PMC article.
-
The Landscape of Micro-Inversions Provide Clues for Population Genetic Analysis of Humans.Interdiscip Sci. 2020 Dec;12(4):499-514. doi: 10.1007/s12539-020-00392-6. Epub 2020 Sep 14. Interdiscip Sci. 2020. PMID: 32929667 Free PMC article.
-
Dynamics of rumen gene expression, microbiome colonization, and their interplay in goats.BMC Genomics. 2021 Apr 21;22(1):288. doi: 10.1186/s12864-021-07595-1. BMC Genomics. 2021. PMID: 33882826 Free PMC article.
-
Expression profile of circular RNA s in TMJ osteoarthritis synovial tissues and potential functions of hsa_circ_0000448 with specific back-spliced junction.Am J Transl Res. 2019 Sep 15;11(9):5357-5374. eCollection 2019. Am J Transl Res. 2019. PMID: 31632516 Free PMC article.
-
Influenza Virus NS1 Protein-RNA Interactome Reveals Intron Targeting.J Virol. 2018 Nov 27;92(24):e01634-18. doi: 10.1128/JVI.01634-18. Print 2018 Dec 15. J Virol. 2018. PMID: 30258002 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

