Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration
- PMID: 26649819
- PMCID: PMC4720437
- DOI: 10.1038/nature16460
Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration
Abstract
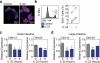
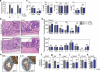
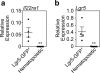
Epithelial regeneration is critical for barrier maintenance and organ function after intestinal injury. The intestinal stem cell (ISC) niche provides Wnt, Notch and epidermal growth factor (EGF) signals supporting Lgr5(+) crypt base columnar ISCs for normal epithelial maintenance. However, little is known about the regulation of the ISC compartment after tissue damage. Using ex vivo organoid cultures, here we show that innate lymphoid cells (ILCs), potent producers of interleukin-22 (IL-22) after intestinal injury, increase the growth of mouse small intestine organoids in an IL-22-dependent fashion. Recombinant IL-22 directly targeted ISCs, augmenting the growth of both mouse and human intestinal organoids, increasing proliferation and promoting ISC expansion. IL-22 induced STAT3 phosphorylation in Lgr5(+) ISCs, and STAT3 was crucial for both organoid formation and IL-22-mediated regeneration. Treatment with IL-22 in vivo after mouse allogeneic bone marrow transplantation enhanced the recovery of ISCs, increased epithelial regeneration and reduced intestinal pathology and mortality from graft-versus-host disease. ATOH1-deficient organoid culture demonstrated that IL-22 induced epithelial regeneration independently of the Paneth cell niche. Our findings reveal a fundamental mechanism by which the immune system is able to support the intestinal epithelium, activating ISCs to promote regeneration.
Figures









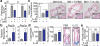
Comment in
-
Regenerative biology: Innate immunity repairs gut lining.Nature. 2015 Dec 24;528(7583):488-9. doi: 10.1038/nature16325. Epub 2015 Dec 9. Nature. 2015. PMID: 26649826 No abstract available.
-
Stem cells: IL-22 activates ISCs for intestinal regeneration.Nat Rev Gastroenterol Hepatol. 2016 Feb;13(2):64. doi: 10.1038/nrgastro.2015.221. Epub 2015 Dec 23. Nat Rev Gastroenterol Hepatol. 2016. PMID: 26695084 No abstract available.
Similar articles
-
ATF3 Sustains IL-22-Induced STAT3 Phosphorylation to Maintain Mucosal Immunity Through Inhibiting Phosphatases.Front Immunol. 2018 Nov 5;9:2522. doi: 10.3389/fimmu.2018.02522. eCollection 2018. Front Immunol. 2018. PMID: 30455690 Free PMC article.
-
IL22 Inhibits Epithelial Stem Cell Expansion in an Ileal Organoid Model.Cell Mol Gastroenterol Hepatol. 2018 Jul 4;7(1):1-17. doi: 10.1016/j.jcmgh.2018.06.008. eCollection 2019. Cell Mol Gastroenterol Hepatol. 2018. PMID: 30364840 Free PMC article.
-
Analysis of Aged Dysfunctional Intestinal Stem Cells.Methods Mol Biol. 2020;2171:41-52. doi: 10.1007/978-1-0716-0747-3_3. Methods Mol Biol. 2020. PMID: 32705634
-
Plasticity of Intestinal Epithelium: Stem Cell Niches and Regulatory Signals.Int J Mol Sci. 2020 Dec 31;22(1):357. doi: 10.3390/ijms22010357. Int J Mol Sci. 2020. PMID: 33396437 Free PMC article. Review.
-
Modeling Intestinal Stem Cell Function with Organoids.Int J Mol Sci. 2021 Oct 9;22(20):10912. doi: 10.3390/ijms222010912. Int J Mol Sci. 2021. PMID: 34681571 Free PMC article. Review.
Cited by
-
Long noncoding RNA uc.230/CUG-binding protein 1 axis sustains intestinal epithelial homeostasis and response to tissue injury.JCI Insight. 2022 Oct 10;7(19):e156612. doi: 10.1172/jci.insight.156612. JCI Insight. 2022. PMID: 36214222 Free PMC article.
-
Krüppel-like factor 5 activates chick intestinal stem cell and promotes mucosal repair after impairment.Cell Cycle. 2023 Oct;22(19):2142-2160. doi: 10.1080/15384101.2023.2278938. Epub 2023 Dec 5. Cell Cycle. 2023. PMID: 37950881 Free PMC article.
-
Anterior Gradient Protein 2 Promotes Mucosal Repair in Pediatric Ulcerative Colitis.Biomed Res Int. 2021 May 18;2021:6483860. doi: 10.1155/2021/6483860. eCollection 2021. Biomed Res Int. 2021. PMID: 34055987 Free PMC article.
-
Pathways regulating intestinal stem cells and potential therapeutic targets for radiation enteropathy.Mol Biomed. 2024 Oct 10;5(1):46. doi: 10.1186/s43556-024-00211-0. Mol Biomed. 2024. PMID: 39388072 Free PMC article. Review.
-
Innate Lymphoid Cells as Regulators of Epithelial Integrity: Therapeutic Implications for Inflammatory Bowel Diseases.Front Med (Lausanne). 2021 Mar 30;8:656745. doi: 10.3389/fmed.2021.656745. eCollection 2021. Front Med (Lausanne). 2021. PMID: 33869257 Free PMC article. Review.
References
-
- Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
-
- Metcalfe C, Kljavin NM, Ybarra R, de Sauvage FJ. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHSN272200900059C/PHS HHS/United States
- R01 AI101406/AI/NIAID NIH HHS/United States
- HHSN272200900059C/AI/NIAID NIH HHS/United States
- R01 HL069929/HL/NHLBI NIH HHS/United States
- P01-CA023766/CA/NCI NIH HHS/United States
- P01 CA023766/CA/NCI NIH HHS/United States
- R01 CA142826/CA/NCI NIH HHS/United States
- K22 CA181280/CA/NCI NIH HHS/United States
- K99 CA176376/CA/NCI NIH HHS/United States
- R01 AI080455/AI/NIAID NIH HHS/United States
- K99-CA176376/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U19 AI116497/AI/NIAID NIH HHS/United States
- R01-HL125571/HL/NHLBI NIH HHS/United States
- R01-AI100288/AI/NIAID NIH HHS/United States
- R01 HL125571/HL/NHLBI NIH HHS/United States
- R01-HL069929/HL/NHLBI NIH HHS/United States
- R01 AI100288/AI/NIAID NIH HHS/United States
- P30-CA008748/CA/NCI NIH HHS/United States
- K08 HL115355/HL/NHLBI NIH HHS/United States
- K08-HL115355/HL/NHLBI NIH HHS/United States
- T32 GM007739/GM/NIGMS NIH HHS/United States
- R01-AI101406/AI/NIAID NIH HHS/United States
- R01-AI080455/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

