An Adenovirus DNA Replication Factor, but Not Incoming Genome Complexes, Targets PML Nuclear Bodies
- PMID: 26608315
- PMCID: PMC4719639
- DOI: 10.1128/JVI.02545-15
An Adenovirus DNA Replication Factor, but Not Incoming Genome Complexes, Targets PML Nuclear Bodies
Abstract
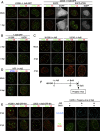
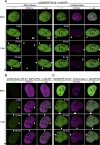
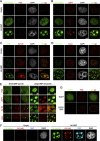
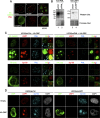
Promyelocytic leukemia protein nuclear bodies (PML-NBs) are subnuclear domains implicated in cellular antiviral responses. Despite the antiviral activity, several nuclear replicating DNA viruses use the domains as deposition sites for the incoming viral genomes and/or as sites for viral DNA replication, suggesting that PML-NBs are functionally relevant during early viral infection to establish productive replication. Although PML-NBs and their components have also been implicated in the adenoviral life cycle, it remains unclear whether incoming adenoviral genome complexes target PML-NBs. Here we show using immunofluorescence and live-cell imaging analyses that incoming adenovirus genome complexes neither localize at nor recruit components of PML-NBs during early phases of infection. We further show that the viral DNA binding protein (DBP), an early expressed viral gene and essential DNA replication factor, independently targets PML-NBs. We show that DBP oligomerization is required to selectively recruit the PML-NB components Sp100 and USP7. Depletion experiments suggest that the absence of one PML-NB component might not affect the recruitment of other components toward DBP oligomers. Thus, our findings suggest a model in which an adenoviral DNA replication factor, but not incoming viral genome complexes, targets and modulates PML-NBs to support a conducive state for viral DNA replication and argue against a generalized concept that PML-NBs target incoming viral genomes.
Importance: The immediate fate upon nuclear delivery of genomes of incoming DNA viruses is largely unclear. Early reports suggested that incoming genomes of herpesviruses are targeted and repressed by PML-NBs immediately upon nuclear import. Genome localization and/or viral DNA replication has also been observed at PML-NBs for other DNA viruses. Thus, it was suggested that PML-NBs may immediately sense and target nuclear viral genomes and hence serve as sites for deposition of incoming viral genomes and/or subsequent viral DNA replication. Here we performed a detailed analyses of the spatiotemporal distribution of incoming adenoviral genome complexes and found, in contrast to the expectation, that an adenoviral DNA replication factor, but not incoming genomes, targets PML-NBs. Thus, our findings may explain why adenoviral genomes could be observed at PML-NBs in earlier reports but argue against a generalized role for PML-NBs in targeting invading viral genomes.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
The interactions between PML nuclear bodies and small and medium size DNA viruses.Virol J. 2023 May 1;20(1):82. doi: 10.1186/s12985-023-02049-4. Virol J. 2023. PMID: 37127643 Free PMC article. Review.
-
PML nuclear body-residing proteins sequentially associate with HPV genome after infectious nuclear delivery.PLoS Pathog. 2019 Feb 25;15(2):e1007590. doi: 10.1371/journal.ppat.1007590. eCollection 2019 Feb. PLoS Pathog. 2019. PMID: 30802273 Free PMC article.
-
Viral DNA Binding Protein SUMOylation Promotes PML Nuclear Body Localization Next to Viral Replication Centers.mBio. 2020 Mar 17;11(2):e00049-20. doi: 10.1128/mBio.00049-20. mBio. 2020. PMID: 32184235 Free PMC article.
-
Sp100 isoform-specific regulation of human adenovirus 5 gene expression.J Virol. 2014 Jun;88(11):6076-92. doi: 10.1128/JVI.00469-14. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623443 Free PMC article.
-
Viruses as hijackers of PML nuclear bodies.Arch Immunol Ther Exp (Warsz). 2003;51(5):295-300. Arch Immunol Ther Exp (Warsz). 2003. PMID: 14626429 Review.
Cited by
-
The interactions between PML nuclear bodies and small and medium size DNA viruses.Virol J. 2023 May 1;20(1):82. doi: 10.1186/s12985-023-02049-4. Virol J. 2023. PMID: 37127643 Free PMC article. Review.
-
Dual signaling via interferon and DNA damage response elicits entrapment by giant PML nuclear bodies.Elife. 2022 Mar 23;11:e73006. doi: 10.7554/eLife.73006. Elife. 2022. PMID: 35319461 Free PMC article.
-
Replication Compartments of DNA Viruses in the Nucleus: Location, Location, Location.Viruses. 2020 Jan 29;12(2):151. doi: 10.3390/v12020151. Viruses. 2020. PMID: 32013091 Free PMC article. Review.
-
A Tale of Usurpation and Subversion: SUMO-Dependent Integrity of Promyelocytic Leukemia Nuclear Bodies at the Crossroad of Infection and Immunity.Front Cell Dev Biol. 2021 Aug 27;9:696234. doi: 10.3389/fcell.2021.696234. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34513832 Free PMC article. Review.
-
Cellular Zinc Finger Protein 622 Hinders Human Adenovirus Lytic Growth and Limits Binding of the Viral pVII Protein to Virus DNA.J Virol. 2019 Jan 17;93(3):e01628-18. doi: 10.1128/JVI.01628-18. Print 2019 Feb 1. J Virol. 2019. PMID: 30429337 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

