Gctf: Real-time CTF determination and correction
- PMID: 26592709
- PMCID: PMC4711343
- DOI: 10.1016/j.jsb.2015.11.003
Gctf: Real-time CTF determination and correction
Abstract
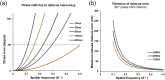
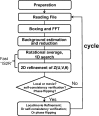
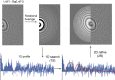
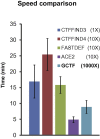
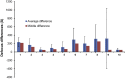
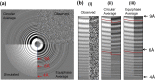
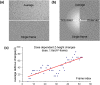
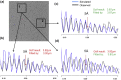
Accurate estimation of the contrast transfer function (CTF) is critical for a near-atomic resolution cryo electron microscopy (cryoEM) reconstruction. Here, a GPU-accelerated computer program, Gctf, for accurate and robust, real-time CTF determination is presented. The main target of Gctf is to maximize the cross-correlation of a simulated CTF with the logarithmic amplitude spectra (LAS) of observed micrographs after background subtraction. Novel approaches in Gctf improve both speed and accuracy. In addition to GPU acceleration (e.g. 10-50×), a fast '1-dimensional search plus 2-dimensional refinement (1S2R)' procedure further speeds up Gctf. Based on the global CTF determination, the local defocus for each particle and for single frames of movies is accurately refined, which improves CTF parameters of all particles for subsequent image processing. Novel diagnosis method using equiphase averaging (EPA) and self-consistency verification procedures have also been implemented in the program for practical use, especially for aims of near-atomic reconstruction. Gctf is an independent program and the outputs can be easily imported into other cryoEM software such as Relion (Scheres, 2012) and Frealign (Grigorieff, 2007). The results from several representative datasets are shown and discussed in this paper.
Keywords: CTF determination; Contrast transfer function; Cryo-electron microscopy; GPU program.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Local defocus estimation in single particle analysis in cryo-electron microscopy.J Struct Biol. 2023 Dec;215(4):108030. doi: 10.1016/j.jsb.2023.108030. Epub 2023 Sep 25. J Struct Biol. 2023. PMID: 37758154
-
A Robust Single-Particle Cryo-Electron Microscopy (cryo-EM) Processing Workflow with cryoSPARC, RELION, and Scipion.J Vis Exp. 2022 Jan 31;(179). doi: 10.3791/63387. J Vis Exp. 2022. PMID: 35104261
-
TomoAlign: A novel approach to correcting sample motion and 3D CTF in CryoET.J Struct Biol. 2021 Dec;213(4):107778. doi: 10.1016/j.jsb.2021.107778. Epub 2021 Aug 18. J Struct Biol. 2021. PMID: 34416376
-
Processing of Structurally Heterogeneous Cryo-EM Data in RELION.Methods Enzymol. 2016;579:125-57. doi: 10.1016/bs.mie.2016.04.012. Epub 2016 May 31. Methods Enzymol. 2016. PMID: 27572726 Review.
-
Frealign: An Exploratory Tool for Single-Particle Cryo-EM.Methods Enzymol. 2016;579:191-226. doi: 10.1016/bs.mie.2016.04.013. Epub 2016 Jun 7. Methods Enzymol. 2016. PMID: 27572728 Free PMC article. Review.
Cited by
-
Characterization of Medusavirus encoded histones reveals nucleosome-like structures and a unique linker histone.Nat Commun. 2024 Oct 23;15(1):9138. doi: 10.1038/s41467-024-53364-5. Nat Commun. 2024. PMID: 39443461 Free PMC article.
-
In vitro structural maturation of an early stage pre-40S particle coupled with U3 snoRNA release and central pseudoknot formation.Nucleic Acids Res. 2022 Nov 11;50(20):11916-11923. doi: 10.1093/nar/gkac910. Nucleic Acids Res. 2022. PMID: 36263816 Free PMC article.
-
Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state.Nat Struct Mol Biol. 2020 Nov;27(11):1009-1016. doi: 10.1038/s41594-020-0481-x. Epub 2020 Aug 24. Nat Struct Mol Biol. 2020. PMID: 32839613 Free PMC article.
-
Structural characterisation of the Chaetomium thermophilum Chl1 helicase.PLoS One. 2021 May 10;16(5):e0251261. doi: 10.1371/journal.pone.0251261. eCollection 2021. PLoS One. 2021. PMID: 33970942 Free PMC article.
-
Molecular basis of inhibition of the amino acid transporter B0AT1 (SLC6A19).Nat Commun. 2024 Aug 22;15(1):7224. doi: 10.1038/s41467-024-51748-1. Nat Commun. 2024. PMID: 39174516 Free PMC article.
References
-
- Bai X.C., McMullan G., Scheres S.H. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 2015;40:49–57. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

