Increased Valency of Conserved-mosaic Vaccines Enhances the Breadth and Depth of Epitope Recognition
- PMID: 26581160
- PMCID: PMC4817818
- DOI: 10.1038/mt.2015.210
Increased Valency of Conserved-mosaic Vaccines Enhances the Breadth and Depth of Epitope Recognition
Abstract
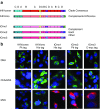
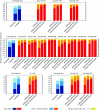
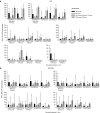
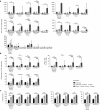
The biggest roadblock in development of effective vaccines against human immunodeficiency virus type 1 (HIV-1) is the virus genetic diversity. For T-cell vaccine, this can be tackled by focusing the vaccine-elicited T-cells on the highly functionally conserved regions of HIV-1 proteins, mutations in which typically cause a replicative fitness loss, and by computing multivalent mosaic proteins, which maximize the coverage of potential 9-mer T-cell epitopes of the input viral sequences. Our first conserved region vaccines HIVconsv employed clade alternating consensus sequences and showed promise in the initial clinical trials in terms of magnitude and breadth of elicited CD8(+) T-cells. Here, monitoring T-cells restricted by HLA-A*02:01 in transgenic mice, we assessed whether or not the tHIVconsv design (HIVconsv with a tissue plasminogen activator leader sequence) benefits from combining with a complementing conserved mosaic immunogen tHIVcmo, and compared the bivalent immunization to that with trivalent conserved mosaic vaccines. A hierarchy of tHIVconsv ≤ tHIVconsv+tHIVcmo < tCmo1+tCmo2+tCmo3 vaccinations for induction of CD8(+) T-cell responses was observed in terms of recognition of tested peptide variants. Thus, our HLA-A*02:01-restricted epitope data concur with previously published mouse and macaque observations and suggest that even conserved region vaccines benefit from oligovalent mosaic design.
Figures

Similar articles
-
Novel, in-natural-infection subdominant HIV-1 CD8+ T-cell epitopes revealed in human recipients of conserved-region T-cell vaccines.PLoS One. 2017 Apr 27;12(4):e0176418. doi: 10.1371/journal.pone.0176418. eCollection 2017. PLoS One. 2017. PMID: 28448594 Free PMC article. Clinical Trial.
-
Novel Conserved-region T-cell Mosaic Vaccine With High Global HIV-1 Coverage Is Recognized by Protective Responses in Untreated Infection.Mol Ther. 2016 Apr;24(4):832-42. doi: 10.1038/mt.2016.3. Epub 2016 Jan 8. Mol Ther. 2016. PMID: 26743582 Free PMC article.
-
Control of HIV-1 replication in vitro by vaccine-induced human CD8(+) T cells through conserved subdominant Pol epitopes.Vaccine. 2016 Feb 24;34(9):1215-24. doi: 10.1016/j.vaccine.2015.12.021. Epub 2016 Jan 16. Vaccine. 2016. PMID: 26784683 Free PMC article.
-
Multiple Approaches for Increasing the Immunogenicity of an Epitope-Based Anti-HIV Vaccine.AIDS Res Hum Retroviruses. 2015 Nov;31(11):1077-88. doi: 10.1089/AID.2015.0101. Epub 2015 Aug 13. AIDS Res Hum Retroviruses. 2015. PMID: 26149745 Review.
-
Aiming for protective T-cell responses: a focus on the first generation conserved-region HIVconsv vaccines in preventive and therapeutic clinical trials.Expert Rev Vaccines. 2019 Oct;18(10):1029-1041. doi: 10.1080/14760584.2019.1675518. Epub 2019 Oct 15. Expert Rev Vaccines. 2019. PMID: 31613649 Review.
Cited by
-
Mosaic H5 Hemagglutinin Provides Broad Humoral and Cellular Immune Responses against Influenza Viruses.J Virol. 2016 Jul 11;90(15):6771-6783. doi: 10.1128/JVI.00730-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27194759 Free PMC article.
-
Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13-19).Lancet. 2018 Jul 21;392(10143):232-243. doi: 10.1016/S0140-6736(18)31364-3. Epub 2018 Jul 6. Lancet. 2018. PMID: 30047376 Free PMC article. Clinical Trial.
-
HIV-1 Conserved Mosaics Delivered by Regimens with Integration-Deficient DC-Targeting Lentiviral Vector Induce Robust T Cells.Mol Ther. 2017 Feb 1;25(2):494-503. doi: 10.1016/j.ymthe.2016.12.004. Mol Ther. 2017. PMID: 28153096 Free PMC article.
-
Learning from HIV-1 to predict the immunogenicity of T cell epitopes in SARS-CoV-2.iScience. 2021 Apr 23;24(4):102311. doi: 10.1016/j.isci.2021.102311. Epub 2021 Mar 15. iScience. 2021. PMID: 33748696 Free PMC article.
-
Polyvalent vaccine approaches to combat HIV-1 diversity.Immunol Rev. 2017 Jan;275(1):230-244. doi: 10.1111/imr.12516. Immunol Rev. 2017. PMID: 28133800 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

