Shared Genetic Signals of Hypoxia Adaptation in Drosophila and in High-Altitude Human Populations
- PMID: 26576852
- PMCID: PMC4866538
- DOI: 10.1093/molbev/msv248
Shared Genetic Signals of Hypoxia Adaptation in Drosophila and in High-Altitude Human Populations
Abstract
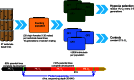
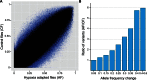
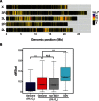
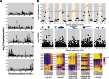
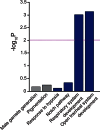
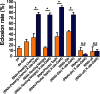
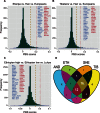
The ability to withstand low oxygen (hypoxia tolerance) is a polygenic and mechanistically conserved trait that has important implications for both human health and evolution. However, little is known about the diversity of genetic mechanisms involved in hypoxia adaptation in evolving populations. We used experimental evolution and whole-genome sequencing in Drosophila melanogaster to investigate the role of natural variation in adaptation to hypoxia. Using a generalized linear mixed model we identified significant allele frequency differences between three independently evolved hypoxia-tolerant populations and normoxic control populations for approximately 3,800 single nucleotide polymorphisms. Around 50% of these variants are clustered in 66 distinct genomic regions. These regions contain genes that are differentially expressed between hypoxia-tolerant and normoxic populations and several of the differentially expressed genes are associated with metabolic processes. Additional genes associated with respiratory and open tracheal system development also show evidence of directional selection. RNAi-mediated knockdown of several candidate genes' expression significantly enhanced survival in severe hypoxia. Using genomewide single nucleotide polymorphism data from four high-altitude human populations-Sherpas, Tibetans, Ethiopians, and Andeans, we found that several human orthologs of the genes under selection in flies are also likely under positive selection in all four high-altitude human populations. Thus, our results indicate that selection for hypoxia tolerance can act on standing genetic variation in similar genes and pathways present in organisms diverged by hundreds of millions of years.
Keywords: adaptation; complex traits; evolution; evolve and resequence; experimental evolution; high-altitude adaptation; hypoxia; polygenic traits; pooled sequencing.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
Similar articles
-
Whole-Genome Resequencing of Experimental Populations Reveals Polygenic Basis of Egg-Size Variation in Drosophila melanogaster.Mol Biol Evol. 2015 Oct;32(10):2616-32. doi: 10.1093/molbev/msv136. Epub 2015 Jun 3. Mol Biol Evol. 2015. PMID: 26044351 Free PMC article.
-
Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data.PLoS Genet. 2010 Sep 9;6(9):e1001116. doi: 10.1371/journal.pgen.1001116. PLoS Genet. 2010. PMID: 20838600 Free PMC article.
-
Genetics of human origin and evolution: high-altitude adaptations.Curr Opin Genet Dev. 2016 Dec;41:8-13. doi: 10.1016/j.gde.2016.06.018. Epub 2016 Aug 6. Curr Opin Genet Dev. 2016. PMID: 27501156 Free PMC article. Review.
-
Identifying positive selection candidate loci for high-altitude adaptation in Andean populations.Hum Genomics. 2009 Dec;4(2):79-90. doi: 10.1186/1479-7364-4-2-79. Hum Genomics. 2009. PMID: 20038496 Free PMC article.
-
Measuring high-altitude adaptation.J Appl Physiol (1985). 2017 Nov 1;123(5):1371-1385. doi: 10.1152/japplphysiol.00321.2017. Epub 2017 Aug 31. J Appl Physiol (1985). 2017. PMID: 28860167 Free PMC article. Review.
Cited by
-
Cross-Species Insights Into Genomic Adaptations to Hypoxia.Front Genet. 2020 Jul 22;11:743. doi: 10.3389/fgene.2020.00743. eCollection 2020. Front Genet. 2020. PMID: 32849780 Free PMC article. Review.
-
Artificial Selection Finds New Hypotheses for the Mechanism of Wolbachia-Mediated Dengue Blocking in Mosquitoes.Front Microbiol. 2020 Jul 7;11:1456. doi: 10.3389/fmicb.2020.01456. eCollection 2020. Front Microbiol. 2020. PMID: 32733407 Free PMC article.
-
The genomic basis of high-elevation adaptation in wild house mice (Mus musculus domesticus) from South America.Genetics. 2022 Feb 4;220(2):iyab226. doi: 10.1093/genetics/iyab226. Genetics. 2022. PMID: 34897431 Free PMC article.
-
Hypoxia delays steroid-induced developmental maturation in Drosophila by suppressing EGF signaling.PLoS Genet. 2024 Apr 26;20(4):e1011232. doi: 10.1371/journal.pgen.1011232. eCollection 2024 Apr. PLoS Genet. 2024. PMID: 38669270 Free PMC article.
-
Multiple across-strain and within-strain QTLs suggest highly complex genetic architecture for hypoxia tolerance in channel catfish.Mol Genet Genomics. 2017 Feb;292(1):63-76. doi: 10.1007/s00438-016-1256-2. Epub 2016 Oct 12. Mol Genet Genomics. 2017. PMID: 27734158
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

