Chromatin remodeling and bivalent histone modifications in embryonic stem cells
- PMID: 26553936
- PMCID: PMC4693513
- DOI: 10.15252/embr.201541011
Chromatin remodeling and bivalent histone modifications in embryonic stem cells
Abstract
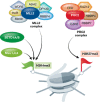
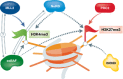
Pluripotent embryonic stem cells (ESCs) are characterized by distinct epigenetic features including a relative enrichment of histone modifications related to active chromatin. Among these is tri-methylation of lysine 4 on histone H3 (H3K4me3). Several thousands of the H3K4me3-enriched promoters in pluripotent cells also contain a repressive histone mark, namely H3K27me3, a situation referred to as "bivalency". While bivalent promoters are not unique to pluripotent cells, they are relatively enriched in these cell types, largely marking developmental and lineage-specific genes which are silent but poised for immediate action. The H3K4me3 and H3K27me3 modifications are catalyzed by lysine methyltransferases which are usually found within, although not entirely limited to, the Trithorax group (TrxG) and Polycomb group (PcG) protein complexes, respectively, but these do not provide selective bivalent specificity. Recent studies highlight the family of ATP-dependent chromatin remodeling proteins as regulators of bivalent domains. Here, we discuss bivalency in general, describe the machineries that catalyze bivalent chromatin domains, and portray the emerging connection between bivalency and the action of different families of chromatin remodelers, namely INO80, esBAF, and NuRD, in pluripotent cells. We posit that chromatin remodeling proteins may enable "bivalent specificity", often selectively acting on, or selectively depleted from, bivalent domains.
Keywords: chromatin; chromatin remodeling; embryonic stem cells; epigenetics; histone modifications.
© 2015 The Authors.
Figures

Similar articles
-
Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant.Development. 2014 Feb;141(3):526-37. doi: 10.1242/dev.102681. Epub 2014 Jan 14. Development. 2014. PMID: 24423662
-
Nucleosomal asymmetry shapes histone mark binding and promotes poising at bivalent domains.Mol Cell. 2025 Feb 6;85(3):471-489.e12. doi: 10.1016/j.molcel.2024.12.002. Epub 2024 Dec 27. Mol Cell. 2025. PMID: 39731917
-
Bivalent promoter hypermethylation in cancer is linked to the H327me3/H3K4me3 ratio in embryonic stem cells.BMC Biol. 2020 Mar 4;18(1):25. doi: 10.1186/s12915-020-0752-3. BMC Biol. 2020. PMID: 32131813 Free PMC article.
-
Bivalent Histone Modifications and Development.Curr Stem Cell Res Ther. 2018;13(2):83-90. doi: 10.2174/1574888X12666170123144743. Curr Stem Cell Res Ther. 2018. PMID: 28117006 Review.
-
Regulation, functions and transmission of bivalent chromatin during mammalian development.Nat Rev Mol Cell Biol. 2023 Jan;24(1):6-26. doi: 10.1038/s41580-022-00518-2. Epub 2022 Aug 26. Nat Rev Mol Cell Biol. 2023. PMID: 36028557 Review.
Cited by
-
LncRNA-Smad7 mediates cross-talk between Nodal/TGF-β and BMP signaling to regulate cell fate determination of pluripotent and multipotent cells.Nucleic Acids Res. 2022 Oct 14;50(18):10526-10543. doi: 10.1093/nar/gkac780. Nucleic Acids Res. 2022. PMID: 36134711 Free PMC article.
-
Genome-wide mapping of histone modifications during axenic growth in two species of Leptosphaeria maculans showing contrasting genomic organization.Chromosome Res. 2021 Jun;29(2):219-236. doi: 10.1007/s10577-021-09658-1. Epub 2021 May 21. Chromosome Res. 2021. PMID: 34018080 Free PMC article.
-
Activation of Galectin-3 (LGALS3) Transcription by Injurious Stimuli in the Liver Is Commonly Mediated by BRG1.Front Cell Dev Biol. 2019 Nov 26;7:310. doi: 10.3389/fcell.2019.00310. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31850346 Free PMC article.
-
Epigenetics in Congenital Heart Disease.J Am Heart Assoc. 2022 Apr 5;11(7):e025163. doi: 10.1161/JAHA.121.025163. Epub 2022 Mar 29. J Am Heart Assoc. 2022. PMID: 35348004 Free PMC article. Review.
-
Chromatin remodeler Activity-Dependent Neuroprotective Protein (ADNP) contributes to syndromic autism.Clin Epigenetics. 2023 Mar 21;15(1):45. doi: 10.1186/s13148-023-01450-8. Clin Epigenetics. 2023. PMID: 36945042 Free PMC article. Review.
References
-
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 - PubMed
-
- Tessarz P, Kouzarides T (2014) Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol 15: 703–708 - PubMed
-
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M et al (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

