Long noncoding RNAs in T lymphocytes
- PMID: 26538526
- PMCID: PMC6608032
- DOI: 10.1189/jlb.1RI0815-389R
Long noncoding RNAs in T lymphocytes
Abstract
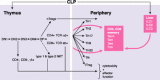
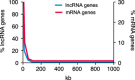
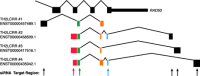
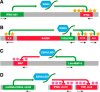
Long noncoding RNAs are recently discovered regulatory RNA molecules that do not code for proteins but influence a vast array of biologic processes. In vertebrates, the number of long noncoding RNA genes is thought to greatly exceed the number of protein-coding genes. It is also thought that long noncoding RNAs drive the biologic complexity observed in vertebrates compared with that in invertebrates. Evidence of this complexity has been found in the T-lymphocyte compartment of the adaptive immune system. In the present review, we describe our current level of understanding of the expression of specific long or large intergenic or intervening long noncoding RNAs during T-lymphocyte development in the thymus and differentiation in the periphery and highlight the mechanisms of action that specific long noncoding RNAs employ to regulate T-lymphocyte function, both in vitro and in vivo.
Keywords: adaptive immunity; epigenetics; lncRNA.
© Society for Leukocyte Biology.
Figures
Similar articles
-
Emerging roles of noncoding RNAs in T cell differentiation and functions in autoimmune diseases.Int Rev Immunol. 2019;38(5):232-245. doi: 10.1080/08830185.2019.1648454. Epub 2019 Aug 14. Int Rev Immunol. 2019. PMID: 31411520 Review.
-
Gene regulation in the immune system by long noncoding RNAs.Nat Immunol. 2017 Aug 22;18(9):962-972. doi: 10.1038/ni.3771. Nat Immunol. 2017. PMID: 28829444 Free PMC article. Review.
-
Long noncoding RNAs in autoimmune diseases.J Biomed Mater Res A. 2019 Feb;107(2):468-475. doi: 10.1002/jbm.a.36562. Epub 2018 Nov 26. J Biomed Mater Res A. 2019. PMID: 30478988 Review.
-
The Emerging Functions of Long Noncoding RNA in Immune Cells: Autoimmune Diseases.J Immunol Res. 2015;2015:848790. doi: 10.1155/2015/848790. Epub 2015 May 18. J Immunol Res. 2015. PMID: 26090502 Free PMC article. Review.
-
Identification of global regulators of T-helper cell lineage specification.Genome Med. 2015 Nov 20;7:122. doi: 10.1186/s13073-015-0237-0. Genome Med. 2015. PMID: 26589177 Free PMC article.
Cited by
-
LncRNAs as Regulators of Autophagy and Drug Resistance in Colorectal Cancer.Front Oncol. 2019 Oct 2;9:1008. doi: 10.3389/fonc.2019.01008. eCollection 2019. Front Oncol. 2019. PMID: 31632922 Free PMC article. Review.
-
The Emerging Role of Epigenetics in Autoimmune Thyroid Diseases.Front Immunol. 2017 Apr 7;8:396. doi: 10.3389/fimmu.2017.00396. eCollection 2017. Front Immunol. 2017. PMID: 28439272 Free PMC article. Review.
-
The emerging role of long non-coding RNA in spinal cord injury.J Cell Mol Med. 2018 Apr;22(4):2055-2061. doi: 10.1111/jcmm.13515. Epub 2018 Feb 1. J Cell Mol Med. 2018. PMID: 29392896 Free PMC article. Review.
-
Multilayer epigenetic analysis reveals novel transcription factor networks in CD8 T cells.Cell Mol Immunol. 2018 Mar;15(3):199-202. doi: 10.1038/cmi.2017.46. Epub 2017 Jul 31. Cell Mol Immunol. 2018. PMID: 28757613 Free PMC article. No abstract available.
-
Inflammatory cells and their non-coding RNAs as targets for treating myocardial infarction.Basic Res Cardiol. 2018 Dec 6;114(1):4. doi: 10.1007/s00395-018-0712-z. Basic Res Cardiol. 2018. PMID: 30523422 Free PMC article. Review.
References
-
- Lander, E.S.L. , Linton, L.M. , Birren, B. , Nusbaum, C. , Zody, M.C. , Baldwin, J. , Devon, K. , Dewar, K. , Doyle, M. , FitzHugh, W. , Funke, R. , Gage, D. , Harris, K. , Heaford, A. , Howland, J. , Kann, L. , Lehoczky, J. , LeVine, R. , McEwan, P. , McKernan, K. , Meldrim, J. , Mesirov, J.P. , Miranda, C. , Morris, W. , Naylor, J. , Raymond, C. , Rosetti, M. , Santos, R. , Sheridan, A. , Sougnez, C. , Stange‐Thomann, Y. , Stojanovic, N. , Subramanian, A. , Wyman, D. , Rogers, J. , Sulston, J. , Ainscough, R. , Beck, S. , Bentley, D. , Burton, J. , Clee, C. , Carter, N. , Coulson, A. , Deadman, R. , Deloukas, P. , Dunham, A. , Dunham, I. , Durbin, R. , French, L. , Grafham, D. , Gregory, S. , Hubbard, T. , Humphray, S. , Hunt, A. , Jones, M. , Lloyd, C. , McMurray, A. , Matthews, L. , Mercer, S. , Milne, S. , Mullikin, J.C. , Mungall, A. , Plumb, R. , Ross, M. , Shownkeen, R. , Sims, S. , Waterston, R.H. , Wilson, R.K. , Hillier, L.W. , McPherson, J.D. , Marra, M.A. , Mardis, E.R. , Fulton, L.A. , Chinwalla, A.T. , Pepin, K.H. , Gish, W.R. , Chissoe, S.L. , Wendl, M.C. , Delehaunty, K.D. , Miner, T.L. , Delehaunty, A. , Kramer, J.B. , Cook, L.L. , Fulton, R.S. , Johnson, D.L. , Minx, P.J. , Clifton, S.W. , Hawkins, T. , Branscomb, E. , Predki, P. , Richardson, P. , Wenning, S. , Slezak, T. , Doggett, N. , Cheng, J.F. , Olsen, A. , Lucas, S. , Elkin, C. , Uberbacher, E. , Frazier, M. , Gibbs, R.A. , Muzny, D.M. , Scherer, S.E. , Bouck, J.B. , Sodergren, E.J. , Worley, K.C. , Rives, C.M. , Gorrell, J.H. , Metzker, M.L. , Naylor, S.L. , Kucherlapati, R.S. , Nelson, D.L. , Weinstock, G.M. , Sakaki, Y. , Fujiyama, A. , Hattori, M. , Yada, T. , Toyoda, A. , Itoh, T. , Kawagoe, C. , Watanabe, H. , Totoki, Y. , Taylor, T. , Weissenbach, J. , Heilig, R. , Saurin, W. , Artiguenave, F. , Brottier, P. , Bruls, T. , Pelletier, E. , Robert, C. , Wincker, P. , Smith, D.R. , Doucette‐Stamm, L. , Rubenfield, M. , Weinstock, K. , Lee, H.M. , Dubois, J. , Rosenthal, A. , Platzer, M. , Nyakatura, G. , Taudien, S. , Rump, A. , Yang, H. , Yu, J. , Wang, J. , Huang, G. , Gu, J. , Hood, L. , Rowen, L. , Madan, A. , Qin, S. , Davis, R.W. , Federspiel, N.A. , Abola, A.P. , Proctor, M.J. , Myers, R.M. , Schmutz, J. , Dickson, M. , Grimwood, J. , Cox, D.R. , Olson, M.V. , Kaul, R. , Raymond, C. , Shimizu, N. , Kawasaki, K. , Minoshima, S. , Evans, G.A. , Athanasiou, M. , Schultz, R. , Roe, B.A. , Chen, F. , Pan, H. , Ramser, J. , Lehrach, H. , Reinhardt, R. , McCombie, W.R. , de la Bastide, M. , Dedhia, N. , Blöcker, H. , Hornischer, K. , Nordsiek, G. , Agarwala, R. , Aravind, L. , Bailey, J.A. , Bateman, A. , Batzoglou, S. , Birney, E. , Bork, P. , Brown, D.G. , Burge, C.B. , Cerutti, L. , Chen, H.C. , Church, D. , Clamp, M. , Copley, R.R. , Doerks, T. , Eddy, S.R. , Eichler, E.E. , Furey, T.S. , Galagan, J. , Gilbert, J.G. , Harmon, C. , Hayashizaki, Y. , Haussler, D. , Hermjakob, H. , Hokamp, K. , Jang, W. , Johnson, L.S. , Jones, T.A. , Kasif, S. , Kaspryzk, A. , Kennedy, S. , Kent, W.J. , Kitts, P. , Koonin, E.V. , Korf, I. , Kulp, D. , Lancet, D. , Lowe, T.M. , McLysaght, A. , Mikkelsen, T. , Moran, J.V. , Mulder, N. , Pollara, V.J. , Ponting, C.P. , Schuler, G. , Schultz, J. , Slater, G. , Smit, A.F. , Stupka, E. , Szustakowski, J. , Thierry‐Mieg, D. , Thierry‐Mieg, J. , Wagner, L. , Wallis, J. , Wheeler, R. , Williams, A. , Wolf, Y.I. , Wolfe, K.H. , Yang, S.P. , Yeh, R.F. , Collins, F. , Guyer, M.S. , Peterson, J. , Felsenfeld, A. , Wetterstrand, K.A. , Patrinos, A. , Morgan, M.J. , de Jong, P. , Catanese, J.J. , Osoegawa, K. , Shizuya, H. , Choi, S. , Chen, Y.J. , Szustakowki, J.; International Human Genome Sequencing Consortium. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921. - PubMed
-
- Venter, J.C. , Adams, M.D. , Myers, E.W. , Li, P.W. , Mural, R.J. , Sutton, G.G. , Smith, H.O. , Yandell, M. , Evans, C.A. , Holt, R.A. , Gocayne, J.D. , Amanatides, P. , Ballew, R.M. , Huson, D.H. , Wortman, J.R. , Zhang, Q. , Kodira, C.D. , Zheng, X.H. , Chen, L. , Skupski, M. , Subramanian, G. , Thomas, P.D. , Zhang, J. , Gabor Miklos, G.L. , Nelson, C. , Broder, S. , Clark, A.G. , Nadeau, J. , McKusick, V.A. , Zinder, N. , Levine, A.J. , Roberts, R.J. , Simon, M. , Slayman, C. , Hunkapiller, M. , Bolanos, R. , Delcher, A. , Dew, I. , Fasulo, D. , Flanigan, M. , Florea, L. , Halpern, A. , Hannenhalli, S. , Kravitz, S. , Levy, S. , Mobarry, C. , Reinert, K. , Remington, K. , Abu‐Threideh, J. , Beasley, E. , Biddick, K. , Bonazzi, V. , Brandon, R. , Cargill, M. , Chandramouliswaran, I. , Charlab, R. , Chaturvedi, K. , Deng, Z. , Di Francesco, V. , Dunn, P. , Eilbeck, K. , Evangelista, C. , Gabrielian, A.E. , Gan, W. , Ge, W. , Gong, F. , Gu, Z. , Guan, P. , Heiman, T.J. , Higgins, M.E. , Ji, R.R. , Ke, Z. , Ketchum, K.A. , Lai, Z. , Lei, Y. , Li, Z. , Li, J. , Liang, Y. , Lin, X. , Lu, F. , Merkulov, G.V. , Milshina, N. , Moore, H.M. , Naik, A.K. , Narayan, V.A. , Neelam, B. , Nusskern, D. , Rusch, D.B. , Salzberg, S. , Shao, W. , Shue, B. , Sun, J. , Wang, Z. , Wang, A. , Wang, X. , Wang, J. , Wei, M. , Wides, R. , Xiao, C. , Yan, C. , Yao, A. , Ye, J. , Zhan, M. , Zhang, W. , Zhang, H. , Zhao, Q. , Zheng, L. , Zhong, F. , Zhong, W. , Zhu, S. , Zhao, S. , Gilbert, D. , Baumhueter, S. , Spier, G. , Carter, C. , Cravchik, A. , Woodage, T. , Ali, F. , An, H. , Awe, A. , Baldwin, D. , Baden, H. , Barnstead, M. , Barrow, I. , Beeson, K. , Busam, D. , Carver, A. , Center, A. , Cheng, M.L. , Curry, L. , Danaher, S. , Davenport, L. , Desilets, R. , Dietz, S. , Dodson, K. , Doup, L. , Ferriera, S. , Garg, N. , Gluecksmann, A. , Hart, B. , Haynes, J. , Haynes, C. , Heiner, C. , Hladun, S. , Hostin, D. , Houck, J. , Howland, T. , Ibegwam, C. , Johnson, J. , Kalush, F. , Kline, L. , Koduru, S. , Love, A. , Mann, F. , May, D. , McCawley, S. , McIntosh, T. , McMullen, I. , Moy, M. , Moy, L. , Murphy, B. , Nelson, K. , Pfannkoch, C. , Pratts, E. , Puri, V. , Qureshi, H. , Reardon, M. , Rodriguez, R. , Rogers, Y.H. , Romblad, D. , Ruhfel, B. , Scott, R. , Sitter, C. , Smallwood, M. , Stewart, E. , Strong, R. , Suh, E. , Thomas, R. , Tint, N.N. , Tse, S. , Vech, C. , Wang, G. , Wetter, J. , Williams, S. , Williams, M. , Windsor, S. , Winn‐Deen, E. , Wolfe, K. , Zaveri, J. , Zaveri, K. , Abril, J.F. , Guigó, R. , Campbell, M.J. , Sjolander, K.V. , Karlak, B. , Kejariwal, A. , Mi, H. , Lazareva, B. , Hatton, T. , Narechania, A. , Diemer, K. , Muruganujan, A. , Guo, N. , Sato, S. , Bafna, V. , Istrail, S. , Lippert, R. , Schwartz, R. , Walenz, B. , Yooseph, S. , Allen, D. , Basu, A. , Baxendale, J. , Blick, L. , Caminha, M. , Carnes‐Stine, J. , Caulk, P. , Chiang, Y.H. , Coyne, M. , Dahlke, C. , Mays, A. , Dombroski, M. , Donnelly, M. , Ely, D. , Esparham, S. , Fosler, C. , Gire, H. , Glanowski, S. , Glasser, K. , Glodek, A. , Gorokhov, M. , Graham, K. , Gropman, B. , Harris, M. , Heil, J. , Henderson, S. , Hoover, J. , Jennings, D. , Jordan, C. , Jordan, J. , Kasha, J. , Kagan, L. , Kraft, C. , Levitsky, A. , Lewis, M. , Liu, X. , Lopez, J. , Ma, D. , Majoros, W. , McDaniel, J. , Murphy, S. , Newman, M. , Nguyen, T. , Nguyen, N. , Nodell, M. , Pan, S. , Peck, J. , Peterson, M. , Rowe, W. , Sanders, R. , Scott, J. , Simpson, M. , Smith, T. , Sprague, A. , Stockwell, T. , Turner, R. , Venter, E. , Wang, M. , Wen, M. , Wu, D. , Wu, M. , Xia, A. , Zandieh, A. , Zhu, X. (2001) The sequence of the human genome. Science 291, 1304–1351. - PubMed
-
- Harrow, J. , Frankish, A. , Gonzalez, J.M. , Tapanari, E. , Diekhans, M. , Kokocinski, F. , Aken, B.L. , Barrell, D. , Zadissa, A. , Searle, S. , Barnes, I. , Bignell, A. , Boychenko, V. , Hunt, T. , Kay, M. , Mukherjee, G. , Rajan, J. , Despacio‐Reyes, G. , Saunders, G. , Steward, C. , Harte, R. , Lin, M. , Howald, C. , Tanzer, A. , Derrien, T. , Chrast, J. , Walters, N. , Balasubramanian, S. , Pei, B. , Tress, M. , Rodriguez, J.M. , Ezkurdia, I. , van Baren, J. , Brent, M. , Haussler, D. , Kellis, M. , Valencia, A. , Reymond, A. , Gerstein, M. , Guigó, R. , Hubbard, T.J. (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

