Trypanosome RNA editing: the complexity of getting U in and taking U out
- PMID: 26522170
- PMCID: PMC4835692
- DOI: 10.1002/wrna.1313
Trypanosome RNA editing: the complexity of getting U in and taking U out
Abstract
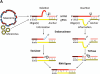
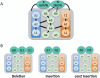
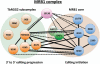
RNA editing, which adds sequence information to RNAs post-transcriptionally, is a widespread phenomenon throughout eukaryotes. The most complex form of this process is the uridine (U) insertion/deletion editing that occurs in the mitochondria of kinetoplastid protists. RNA editing in these flagellates is specified by trans-acting guide RNAs and entails the insertion of hundreds and deletion of dozens of U residues from mitochondrial RNAs to produce mature, translatable mRNAs. An emerging model indicates that the machinery required for trypanosome RNA editing is much more complicated than previously appreciated. A family of RNA editing core complexes (RECCs), which contain the required enzymes and several structural proteins, catalyze cycles of U insertion and deletion. A second, dynamic multiprotein complex, the Mitochondrial RNA Binding 1 (MRB1) complex, has recently come to light as another essential component of the trypanosome RNA editing machinery. MRB1 likely serves as the platform for kinetoplastid RNA editing, and plays critical roles in RNA utilization and editing processivity. MRB1 also appears to act as a hub for coordination of RNA editing with additional mitochondrial RNA processing events. This review highlights the current knowledge regarding the complex molecular machinery involved in trypanosome RNA editing. WIREs RNA 2016, 7:33-51. doi: 10.1002/wrna.1313 For further resources related to this article, please visit the WIREs website.
© 2015 Wiley Periodicals, Inc.
Figures


Similar articles
-
U-Insertion/Deletion mRNA-Editing Holoenzyme: Definition in Sight.Trends Parasitol. 2016 Feb;32(2):144-156. doi: 10.1016/j.pt.2015.10.004. Epub 2015 Nov 10. Trends Parasitol. 2016. PMID: 26572691 Free PMC article. Review.
-
Integrity of the core mitochondrial RNA-binding complex 1 is vital for trypanosome RNA editing.RNA. 2015 Dec;21(12):2088-102. doi: 10.1261/rna.052340.115. Epub 2015 Oct 7. RNA. 2015. PMID: 26447184 Free PMC article.
-
Dual core processing: MRB1 is an emerging kinetoplast RNA editing complex.Trends Parasitol. 2013 Feb;29(2):91-9. doi: 10.1016/j.pt.2012.11.005. Epub 2013 Jan 8. Trends Parasitol. 2013. PMID: 23305619 Free PMC article. Review.
-
Architecture of the trypanosome RNA editing accessory complex, MRB1.Nucleic Acids Res. 2012 Jul;40(12):5637-50. doi: 10.1093/nar/gks211. Epub 2012 Mar 6. Nucleic Acids Res. 2012. PMID: 22396527 Free PMC article.
-
A core MRB1 complex component is indispensable for RNA editing in insect and human infective stages of Trypanosoma brucei.PLoS One. 2013 Oct 18;8(10):e78015. doi: 10.1371/journal.pone.0078015. eCollection 2013. PLoS One. 2013. PMID: 24250748 Free PMC article.
Cited by
-
Poly(A) binding KPAF4/5 complex stabilizes kinetoplast mRNAs in Trypanosoma brucei.Nucleic Acids Res. 2020 Sep 4;48(15):8645-8662. doi: 10.1093/nar/gkaa575. Nucleic Acids Res. 2020. PMID: 32614436 Free PMC article.
-
Post-transcriptional mending of gene sequences: Looking under the hood of mitochondrial gene expression in diplonemids.RNA Biol. 2016 Dec;13(12):1204-1211. doi: 10.1080/15476286.2016.1240143. Epub 2016 Oct 7. RNA Biol. 2016. PMID: 27715490 Free PMC article. Review.
-
Oxidative Phosphorylation Is Required for Powering Motility and Development of the Sleeping Sickness Parasite Trypanosoma brucei in the Tsetse Fly Vector.mBio. 2022 Feb 22;13(1):e0235721. doi: 10.1128/mbio.02357-21. Epub 2022 Jan 11. mBio. 2022. PMID: 35012336 Free PMC article.
-
The large repertoire of 2'-O-methylation guided by C/D snoRNAs on Trypanosoma brucei rRNA.RNA Biol. 2020 Jul;17(7):1018-1039. doi: 10.1080/15476286.2020.1750842. Epub 2020 Apr 21. RNA Biol. 2020. PMID: 32250712 Free PMC article.
-
Pseudouridines on Trypanosoma brucei spliceosomal small nuclear RNAs and their implication for RNA and protein interactions.Nucleic Acids Res. 2019 Aug 22;47(14):7633-7647. doi: 10.1093/nar/gkz477. Nucleic Acids Res. 2019. PMID: 31147702 Free PMC article.
References
-
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. - PubMed
-
- Benne R, Van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, Tromp MC. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. - PubMed
-
- Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. - PubMed
-
- Maier RM, Zeltz P, Kossel H, Bonnard G, Gualberto JM, Grienenberger JM. RNA editing in plant mitochondria and chloroplasts. Plant Mol Biol. 1996;32:343–365. - PubMed
-
- Maas S, Rich A, Nishikura K. A-to-I RNA editing: recent news and residual mysteries. J Biol Chem. 2003;278:1391–1394. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

