How specific is CRISPR/Cas9 really?
- PMID: 26517564
- PMCID: PMC4684463
- DOI: 10.1016/j.cbpa.2015.10.001
How specific is CRISPR/Cas9 really?
Abstract
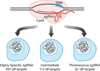
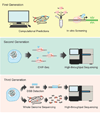
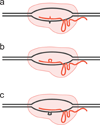
The specificity of RNA-guided nucleases has gathered considerable interest as they become broadly applied to basic research and therapeutic development. Reports of the simple generation of animal models and genome engineering of cells raised questions about targeting precision. Conflicting early reports led the field to believe that CRISPR/Cas9 system was promiscuous, leading to a variety of strategies for improving specificity and increasingly sensitive methods to detect off-target events. However, other studies have suggested that CRISPR/Cas9 is a highly specific genome-editing tool. This review will focus on deciphering and interpreting these seemingly opposing claims.
Copyright © 2015 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
Similar articles
-
Improving CRISPR-Cas9 On-Target Specificity.Curr Issues Mol Biol. 2018;26:65-80. doi: 10.21775/cimb.026.065. Epub 2017 Sep 7. Curr Issues Mol Biol. 2018. PMID: 28879857 Review.
-
Battling CRISPR-Cas9 off-target genome editing.Cell Biol Toxicol. 2019 Oct;35(5):403-406. doi: 10.1007/s10565-019-09485-5. Epub 2019 Jul 16. Cell Biol Toxicol. 2019. PMID: 31313008 No abstract available.
-
Minimizing off-target effects in CRISPR-Cas9 genome editing.Cell Biol Toxicol. 2019 Oct;35(5):399-401. doi: 10.1007/s10565-019-09486-4. Epub 2019 Jul 17. Cell Biol Toxicol. 2019. PMID: 31317359 Free PMC article. No abstract available.
-
CRISPR Mediated Genome Engineering and its Application in Industry.Curr Issues Mol Biol. 2018;26:81-92. doi: 10.21775/cimb.026.081. Epub 2017 Sep 7. Curr Issues Mol Biol. 2018. PMID: 28879858 Review.
-
Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells.Nat Biotechnol. 2015 Sep;33(9):985-989. doi: 10.1038/nbt.3290. Epub 2015 Jun 29. Nat Biotechnol. 2015. PMID: 26121415 Free PMC article.
Cited by
-
Efficient gene correction of an aberrant splice site in β-thalassaemia iPSCs by CRISPR/Cas9 and single-strand oligodeoxynucleotides.J Cell Mol Med. 2019 Dec;23(12):8046-8057. doi: 10.1111/jcmm.14669. Epub 2019 Oct 21. J Cell Mol Med. 2019. PMID: 31631510 Free PMC article.
-
Gene editing with CRISPR-Cas12a guides possessing ribose-modified pseudoknot handles.Nat Commun. 2021 Nov 15;12(1):6591. doi: 10.1038/s41467-021-26989-z. Nat Commun. 2021. PMID: 34782635 Free PMC article.
-
Cornerstones of CRISPR-Cas in drug discovery and therapy.Nat Rev Drug Discov. 2017 Feb;16(2):89-100. doi: 10.1038/nrd.2016.238. Epub 2016 Dec 23. Nat Rev Drug Discov. 2017. PMID: 28008168 Free PMC article. Review.
-
Using Synthetically Engineered Guide RNAs to Enhance CRISPR Genome Editing Systems in Mammalian Cells.Front Genome Ed. 2021 Jan 28;2:617910. doi: 10.3389/fgeed.2020.617910. eCollection 2020. Front Genome Ed. 2021. PMID: 34713240 Free PMC article. Review.
-
Sensing the DNA-mismatch tolerance of catalytically inactive Cas9 via barcoded DNA nanostructures in solid-state nanopores.Nat Biomed Eng. 2024 Mar;8(3):325-334. doi: 10.1038/s41551-023-01078-2. Epub 2023 Aug 7. Nat Biomed Eng. 2024. PMID: 37550424 Free PMC article.
References
-
-
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013;31(9):822–826. First large study to demonstrate that some sgRNA promote promiscous off-target clevage events, with some off-target sites used as much or more often than the on-target site. However, other sgRNAs in this study showed no off-target activity.
-
-
-
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31(9):827–832. Pulbished in the same issue as Fu et al., this study found only intermediate levels of off-target activity, presenting CRISPR/Cas as fairly specific.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

