MAS C-Terminal Tail Interacting Proteins Identified by Mass Spectrometry- Based Proteomic Approach
- PMID: 26484771
- PMCID: PMC4618059
- DOI: 10.1371/journal.pone.0140872
MAS C-Terminal Tail Interacting Proteins Identified by Mass Spectrometry- Based Proteomic Approach
Abstract
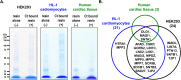
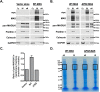
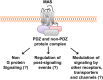
Propagation of signals from G protein-coupled receptors (GPCRs) in cells is primarily mediated by protein-protein interactions. MAS is a GPCR that was initially discovered as an oncogene and is now known to play an important role in cardiovascular physiology. Current literature suggests that MAS interacts with common heterotrimeric G-proteins, but MAS interaction with proteins which might mediate G protein-independent or atypical signaling is unknown. In this study we hypothesized that MAS C-terminal tail (Ct) is a major determinant of receptor-scaffold protein interactions mediating MAS signaling. Mass-spectrometry based proteomic analysis was used to comprehensively identify the proteins that interact with MAS Ct comprising the PDZ-binding motif (PDZ-BM). We identified both PDZ and non-PDZ proteins from human embryonic kidney cell line, mouse atrial cardiomyocyte cell line and human heart tissue to interact specifically with MAS Ct. For the first time our study provides a panel of PDZ and other proteins that potentially interact with MAS with high significance. A 'cardiac-specific finger print' of MAS interacting PDZ proteins was identified which includes DLG1, MAGI1 and SNTA. Cell based experiments with wild-type and mutant MAS lacking the PDZ-BM validated MAS interaction with PDZ proteins DLG1 and TJP2. Bioinformatics analysis suggested well-known multi-protein scaffold complexes involved in nitric oxide signaling (NOS), cell-cell signaling of neuromuscular junctions, synapses and epithelial cells. Majority of these protein hits were predicted to be part of disease categories comprising cancers and malignant tumors. We propose a 'MAS-signalosome' model to stimulate further research in understanding the molecular mechanism of MAS function. Identifying hierarchy of interactions of 'signalosome' components with MAS will be a necessary step in future to fully understand the physiological and pathological functions of this enigmatic receptor.
Conflict of interest statement
Figures



Similar articles
-
PDZ Protein Regulation of G Protein-Coupled Receptor Trafficking and Signaling Pathways.Mol Pharmacol. 2015 Oct;88(4):624-39. doi: 10.1124/mol.115.098509. Epub 2015 Mar 25. Mol Pharmacol. 2015. PMID: 25808930 Review.
-
MAGI proteins can differentially regulate the signaling pathways of 5-HT2AR by enhancing receptor trafficking and PLC recruitment.Cell Signal. 2018 Jul;47:109-121. doi: 10.1016/j.cellsig.2018.03.016. Epub 2018 Apr 3. Cell Signal. 2018. PMID: 29625175
-
Protein kinase Cα promotes cell migration through a PDZ-dependent interaction with its novel substrate discs large homolog 1 (DLG1).J Biol Chem. 2011 Dec 16;286(50):43559-68. doi: 10.1074/jbc.M111.294603. Epub 2011 Oct 25. J Biol Chem. 2011. PMID: 22027822 Free PMC article.
-
The golgi-associated PDZ domain protein PIST/GOPC stabilizes the β1-adrenergic receptor in intracellular compartments after internalization.J Biol Chem. 2015 Mar 6;290(10):6120-9. doi: 10.1074/jbc.M114.605725. Epub 2015 Jan 22. J Biol Chem. 2015. PMID: 25614626 Free PMC article.
-
Subtype-specific roles of phospholipase C-β via differential interactions with PDZ domain proteins.Adv Enzyme Regul. 2011;51(1):138-51. doi: 10.1016/j.advenzreg.2010.10.004. Epub 2010 Oct 28. Adv Enzyme Regul. 2011. PMID: 21035486 Review.
Cited by
-
Connective tissue growth factor dependent collagen gene expression induced by MAS agonist AR234960 in human cardiac fibroblasts.PLoS One. 2017 Dec 29;12(12):e0190217. doi: 10.1371/journal.pone.0190217. eCollection 2017. PLoS One. 2017. PMID: 29287092 Free PMC article.
-
Significance of angiotensin 1-7 coupling with MAS1 receptor and other GPCRs to the renin-angiotensin system: IUPHAR Review 22.Br J Pharmacol. 2017 May;174(9):737-753. doi: 10.1111/bph.13742. Epub 2017 Mar 9. Br J Pharmacol. 2017. PMID: 28194766 Free PMC article. Review.
-
Participation of Gαi-Adenylate Cyclase and ERK1/2 in Mas Receptor Signaling Pathways.Front Pharmacol. 2019 Feb 22;10:146. doi: 10.3389/fphar.2019.00146. eCollection 2019. Front Pharmacol. 2019. PMID: 30853914 Free PMC article.
-
Integrative proteomics and phosphoproteomics in pulmonary arterial hypertension.Sci Rep. 2019 Dec 9;9(1):18623. doi: 10.1038/s41598-019-55053-6. Sci Rep. 2019. PMID: 31819116 Free PMC article.
-
High-mobility group box 1 protein, angiotensins, ACE2, and target organ damage.J Mol Med (Berl). 2016 Jan;94(1):1-3. doi: 10.1007/s00109-015-1372-1. J Mol Med (Berl). 2016. PMID: 26658521 Free PMC article. No abstract available.
References
-
- Young D, Waitches G, Birchmeier C, Fasano O, Wigler M. Isolation and characterization of a new cellular oncogene encoding a protein with multiple potential transmembrane domains. Cell. 1986;45(5):711–9. Epub 1986/06/06. doi: 0092-8674(86)90785-3 [pii]. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

