Mapping asthma-associated variants in admixed populations
- PMID: 26483834
- PMCID: PMC4586512
- DOI: 10.3389/fgene.2015.00292
Mapping asthma-associated variants in admixed populations
Abstract
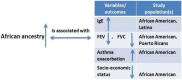
Admixed populations arise when two or more previously isolated populations interbreed. Mapping asthma susceptibility loci in an admixed population using admixture mapping (AM) involves screening the genome of individuals of mixed ancestry for chromosomal regions that have a higher frequency of alleles from a parental population with higher asthma risk as compared with parental population with lower asthma risk. AM takes advantage of the admixture created in populations of mixed ancestry to identify genomic regions where an association exists between genetic ancestry and asthma (in contrast to between the genotype of the marker and asthma). The theory behind AM is that chromosomal segments of affected individuals contain a significantly higher-than-average proportion of alleles from the high-risk parental population and thus are more likely to harbor disease-associated loci. Criteria to evaluate the applicability of AM as a gene mapping approach include: (1) the prevalence of the disease differences in ancestral populations from which the admixed population was formed; (2) a measurable difference in disease-causing alleles between the parental populations; (3) reduced linkage disequilibrium (LD) between unlinked loci across chromosomes and strong LD between neighboring loci; (4) a set of markers with noticeable allele-frequency differences between parental populations that contributes to the admixed population (single nucleotide polymorphisms (SNPs) are the markers of choice because they are abundant, stable, relatively cheap to genotype, and informative with regard to the LD structure of chromosomal segments); and (5) there is an understanding of the extent of segmental chromosomal admixtures and their interactions with environmental factors. Although genome-wide association studies have contributed greatly to our understanding of the genetic components of asthma, the large and increasing degree of admixture in populations across the world create many challenges for further efforts to map disease-causing genes. This review, summarizes the historical context of admixed populations and AM, and considers current opportunities to use AM to map asthma genes. In addition, we provide an overview of the potential limitations and future directions of AM in biomedical research, including joint admixture and association mapping for asthma and asthma-related disorders.
Keywords: admixed population; admixture mapping (AM); ancestry-informative markers (AIMs); asthma; genetic ancestry; genome-wide association study (GWAS); next-generation sequencing (NGS); socio-environmental risk factors.
Figures


Similar articles
-
Genome-wide Association Identifies Novel Etiological Insights Associated with Parkinson's Disease in African and African Admixed Populations.medRxiv [Preprint]. 2023 May 7:2023.05.05.23289529. doi: 10.1101/2023.05.05.23289529. medRxiv. 2023. Update in: Lancet Neurol. 2023 Nov;22(11):1015-1025. doi: 10.1016/S1474-4422(23)00283-1 PMID: 37398408 Free PMC article. Updated. Preprint.
-
Enhanced statistical tests for GWAS in admixed populations: assessment using African Americans from CARe and a Breast Cancer Consortium.PLoS Genet. 2011 Apr;7(4):e1001371. doi: 10.1371/journal.pgen.1001371. Epub 2011 Apr 21. PLoS Genet. 2011. PMID: 21541012 Free PMC article.
-
MI-MAAP: marker informativeness for multi-ancestry admixed populations.BMC Bioinformatics. 2020 Apr 3;21(1):131. doi: 10.1186/s12859-020-3462-5. BMC Bioinformatics. 2020. PMID: 32245404 Free PMC article.
-
Genetic Ancestry Inference and Its Application for the Genetic Mapping of Human Diseases.Int J Mol Sci. 2021 Jun 28;22(13):6962. doi: 10.3390/ijms22136962. Int J Mol Sci. 2021. PMID: 34203440 Free PMC article. Review.
-
Admixture mapping as a gene discovery approach for complex human traits and diseases.Curr Hypertens Rep. 2005 Feb;7(1):31-7. doi: 10.1007/s11906-005-0052-x. Curr Hypertens Rep. 2005. PMID: 15683584 Review.
Cited by
-
Genetic ancestry plays a central role in population pharmacogenomics.Commun Biol. 2021 Feb 5;4(1):171. doi: 10.1038/s42003-021-01681-6. Commun Biol. 2021. PMID: 33547344 Free PMC article.
-
Genetic ancestry differences in pediatric asthma readmission are mediated by socioenvironmental factors.J Allergy Clin Immunol. 2021 Nov;148(5):1210-1218.e4. doi: 10.1016/j.jaci.2021.05.046. Epub 2021 Jul 1. J Allergy Clin Immunol. 2021. PMID: 34217757 Free PMC article.
-
Genetics of Food Allergy.Immunol Allergy Clin North Am. 2021 May;41(2):301-319. doi: 10.1016/j.iac.2021.01.010. Epub 2021 Mar 26. Immunol Allergy Clin North Am. 2021. PMID: 33863485 Free PMC article. Review.
-
Genome-wide admixture and association analysis identifies African ancestry-specific risk loci of eosinophilic esophagitis in African Americans.J Allergy Clin Immunol. 2023 May;151(5):1337-1350. doi: 10.1016/j.jaci.2022.09.040. Epub 2022 Nov 15. J Allergy Clin Immunol. 2023. PMID: 36400179 Free PMC article.
-
Genome-wide association study of asthma, total IgE, and lung function in a cohort of Peruvian children.J Allergy Clin Immunol. 2021 Dec;148(6):1493-1504. doi: 10.1016/j.jaci.2021.02.035. Epub 2021 Mar 10. J Allergy Clin Immunol. 2021. PMID: 33713768 Free PMC article.
References
-
- Akinbami L. J., Moorman J. E., Bailey C., Zahran H. S., King M., Johnson C. A., et al. (2012). Trends in asthma prevalence, health care use, and mortality in the United States, 2001-2010. NCHS Data Brief 94, 1–8. Available online at: http://www.cdc.gov/nchs/data/databriefs/db94.htm - PubMed
-
- Akinbami L. J., Moorman J. E., Liu X. (2011). Asthma prevalence, health care use, and mortality: United States, 2005-2009. Natl. Health Stat. Report 32, 1–14. Available online at: http://www.cdc.gov/nchs/data/nhsr/nhsr032.pdf - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

