Identification of the PhoB Regulon and Role of PhoU in the Phosphate Starvation Response of Caulobacter crescentus
- PMID: 26483520
- PMCID: PMC4686198
- DOI: 10.1128/JB.00658-15
Identification of the PhoB Regulon and Role of PhoU in the Phosphate Starvation Response of Caulobacter crescentus
Abstract
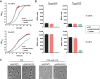
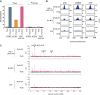
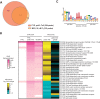
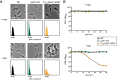
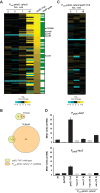
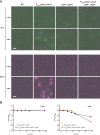
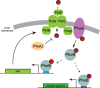
An ability to sense and respond to changes in extracellular phosphate is critical for the survival of most bacteria. For Caulobacter crescentus, which typically lives in phosphate-limited environments, this process is especially crucial. Like many bacteria, Caulobacter responds to phosphate limitation through a conserved two-component signaling pathway called PhoR-PhoB, but the direct regulon of PhoB in this organism is unknown. Here we used chromatin immunoprecipitation-DNA sequencing (ChIP-Seq) to map the global binding patterns of the phosphate-responsive transcriptional regulator PhoB under phosphate-limited and -replete conditions. Combined with genome-wide expression profiling, our work demonstrates that PhoB is induced to regulate nearly 50 genes under phosphate-starved conditions. The PhoB regulon is comprised primarily of genes known or predicted to help Caulobacter scavenge for and import inorganic phosphate, including 15 different membrane transporters. We also investigated the regulatory role of PhoU, a widely conserved protein proposed to coordinate phosphate import with expression of the PhoB regulon by directly modulating the histidine kinase PhoR. However, our studies show that it likely does not play such a role in Caulobacter, as PhoU depletion has no significant effect on PhoB-dependent gene expression. Instead, cells lacking PhoU exhibit striking accumulation of large polyphosphate granules, suggesting that PhoU participates in controlling intracellular phosphate metabolism.
Importance: The transcription factor PhoB is widely conserved throughout the bacterial kingdom, where it helps organisms respond to phosphate limitation by driving the expression of a battery of genes. Most of what is known about PhoB and its target genes is derived from studies of Escherichia coli. Our work documents the PhoB regulon in Caulobacter crescentus, and comparison to the regulon in E. coli reveals significant differences, highlighting the evolutionary plasticity of transcriptional responses driven by highly conserved transcription factors. We also demonstrated that the conserved protein PhoU, which is implicated in bacterial persistence, does not regulate PhoB activity, as previously suggested. Instead, our results favor a model in which PhoU affects intracellular phosphate accumulation, possibly through the high-affinity phosphate transporter.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
PhoU Allows Rapid Adaptation to High Phosphate Concentrations by Modulating PstSCAB Transport Rate in Sinorhizobium meliloti.J Bacteriol. 2017 Aug 22;199(18):e00143-17. doi: 10.1128/JB.00143-17. Print 2017 Sep 15. J Bacteriol. 2017. PMID: 28416708 Free PMC article.
-
Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon.J Bacteriol. 1998 Mar;180(5):1277-86. doi: 10.1128/JB.180.5.1277-1286.1998. J Bacteriol. 1998. PMID: 9495769 Free PMC article.
-
Transcript and protein level analyses of the interactions among PhoB, PhoR, PhoU and CreC in response to phosphate starvation in Escherichia coli.FEMS Microbiol Lett. 2007 Dec;277(2):254-9. doi: 10.1111/j.1574-6968.2007.00965.x. FEMS Microbiol Lett. 2007. PMID: 18031348
-
The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis.FEMS Microbiol Rev. 2008 May;32(3):461-73. doi: 10.1111/j.1574-6976.2008.00101.x. Epub 2008 Jan 29. FEMS Microbiol Rev. 2008. PMID: 18248418 Review.
-
Gene regulation by phosphate in enteric bacteria.J Cell Biochem. 1993 Jan;51(1):47-54. doi: 10.1002/jcb.240510110. J Cell Biochem. 1993. PMID: 8432742 Review.
Cited by
-
Activation of the PhoPR-Mediated Response to Phosphate Limitation Is Regulated by Wall Teichoic Acid Metabolism in Bacillus subtilis.Front Microbiol. 2018 Nov 6;9:2678. doi: 10.3389/fmicb.2018.02678. eCollection 2018. Front Microbiol. 2018. PMID: 30459743 Free PMC article. Review.
-
Quantitative Kinetic Analyses of Shutting Off a Two-Component System.mBio. 2017 May 16;8(3):e00412-17. doi: 10.1128/mBio.00412-17. mBio. 2017. PMID: 28512092 Free PMC article.
-
Microbial river-to-sea continuum: gradients in benthic and planktonic diversity, osmoregulation and nutrient cycling.Microbiome. 2021 Sep 20;9(1):190. doi: 10.1186/s40168-021-01145-3. Microbiome. 2021. PMID: 34544488 Free PMC article.
-
Chemical Reaction Models in Synthetic Promoter Design in Bacteria.Methods Mol Biol. 2024;2844:3-31. doi: 10.1007/978-1-0716-4063-0_1. Methods Mol Biol. 2024. PMID: 39068329
-
Genome-wide mapping of the Escherichia coli PhoB regulon reveals many transcriptionally inert, intragenic binding sites.bioRxiv [Preprint]. 2023 Feb 7:2023.02.07.527549. doi: 10.1101/2023.02.07.527549. bioRxiv. 2023. Update in: mBio. 2023 Jun 27;14(3):e0253522. doi: 10.1128/mbio.02535-22 PMID: 36798257 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

