Identification of Two Novel Members of the Tentative Genus Wukipolyomavirus in Wild Rodents
- PMID: 26474048
- PMCID: PMC4608572
- DOI: 10.1371/journal.pone.0140916
Identification of Two Novel Members of the Tentative Genus Wukipolyomavirus in Wild Rodents
Abstract
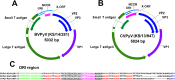
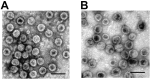
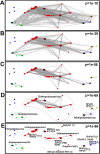
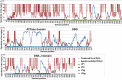
Two novel polyomaviruses (PyVs) were identified in kidney and chest-cavity fluid samples of wild bank voles (Myodes glareolus) and common voles (Microtus arvalis) collected in Germany. All cloned and sequenced genomes had the typical PyV genome organization, including putative open reading frames for early regulatory proteins large T antigen and small T antigen on one strand and for structural late proteins (VP1, VP2 and VP3) on the other strand. Virus-like particles (VLPs) were generated by yeast expression of the VP1 protein of both PyVs. VLP-based ELISA and large T-antigen sequence-targeted polymerase-chain reaction investigations demonstrated signs of infection of these novel PyVs in about 42% of bank voles and 18% of common voles. In most cases only viral DNA, but not VP1-specific antibodies were detected. In additional animals exclusively VP1-specific antibodies, but no viral DNA was detected, indicative for virus clearance. Phylogenetic and clustering analysis including all known PyV genomes placed novel bank vole and common vole PyVs amongst members of the tentative Wukipolymavirus genus. The other known four rodent PyVs, Murine PyV and Hamster PyV, and Murine pneumotropic virus and Mastomys PyV belong to different phylogenetic clades, tentatively named Orthopolyomavirus I and Orthopolyomavirus II, respectively. In conclusion, the finding of novel vole-borne PyVs may suggest an evolutionary origin of ancient wukipolyomaviruses in rodents and may offer the possibility to develop a vole-based animal model for human wukipolyomaviruses.
Conflict of interest statement
Figures

Similar articles
-
Hamster Polyomavirus Research: Past, Present, and Future.Viruses. 2021 May 13;13(5):907. doi: 10.3390/v13050907. Viruses. 2021. PMID: 34068409 Free PMC article. Review.
-
Genomic characterization of two novel polyomaviruses in Brazilian insectivorous bats.Arch Virol. 2015 Jul;160(7):1831-6. doi: 10.1007/s00705-015-2447-6. Epub 2015 May 12. Arch Virol. 2015. PMID: 25963124 Free PMC article.
-
Discovery of African bat polyomaviruses and infrequent recombination in the large T antigen in the Polyomaviridae.J Gen Virol. 2017 Apr;98(4):726-738. doi: 10.1099/jgv.0.000737. Epub 2017 Apr 22. J Gen Virol. 2017. PMID: 28430100
-
Identification of a novel polyomavirus from vervet monkeys in Zambia.J Gen Virol. 2013 Jun;94(Pt 6):1357-1364. doi: 10.1099/vir.0.050740-0. Epub 2013 Feb 20. J Gen Virol. 2013. PMID: 23426354
-
[Epidemiological and basic research activity targeting polyomaviruses].Uirusu. 2014;64(1):25-34. doi: 10.2222/jsv.64.25. Uirusu. 2014. PMID: 25765977 Review. Japanese.
Cited by
-
Isolation and characterization of new Puumala orthohantavirus strains from Germany.Virus Genes. 2020 Aug;56(4):448-460. doi: 10.1007/s11262-020-01755-3. Epub 2020 Apr 23. Virus Genes. 2020. PMID: 32328924 Free PMC article.
-
A novel polyomavirus from the nasal cavity of a giant panda (Ailuropoda melanoleuca).Virol J. 2017 Oct 27;14(1):207. doi: 10.1186/s12985-017-0867-5. Virol J. 2017. PMID: 29078783 Free PMC article.
-
Viral Diversity of House Mice in New York City.mBio. 2018 Apr 17;9(2):e01354-17. doi: 10.1128/mBio.01354-17. mBio. 2018. PMID: 29666290 Free PMC article.
-
The Ancient Evolutionary History of Polyomaviruses.PLoS Pathog. 2016 Apr 19;12(4):e1005574. doi: 10.1371/journal.ppat.1005574. eCollection 2016 Apr. PLoS Pathog. 2016. PMID: 27093155 Free PMC article.
-
Immunogenic Properties and Antigenic Similarity of Virus-like Particles Derived from Human Polyomaviruses.Int J Mol Sci. 2023 Mar 3;24(5):4907. doi: 10.3390/ijms24054907. Int J Mol Sci. 2023. PMID: 36902338 Free PMC article.
References
-
- Imperiale MJ, Major EO. Polyomaviruses In: Knipe DM, Howley PM, Griffen DE, Lamb RA, Martin RA, Roizman B, Straus SE, editors. Fields virology, 4th edn. Lippincott Williams and Wilkins, Philadelphia; 2007. pp. 2263–2298.
-
- Ahuja D, Saenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24:7729–7745. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

