The Role of MicroRNAs as Predictors of Response to Tamoxifen Treatment in Breast Cancer Patients
- PMID: 26473850
- PMCID: PMC4632748
- DOI: 10.3390/ijms161024243
The Role of MicroRNAs as Predictors of Response to Tamoxifen Treatment in Breast Cancer Patients
Abstract
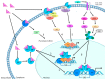
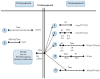
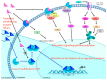
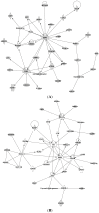
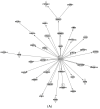
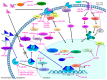
Endocrine therapy is a key treatment strategy to control or eradicate hormone-responsive breast cancer. However, resistance to endocrine therapy leads to breast cancer relapse. The recent extension of adjuvant tamoxifen treatment up to 10 years actualizes the need for identifying biological markers that may be used to monitor predictors of treatment response. MicroRNAs are promising biomarkers that may fill the gap between preclinical knowledge and clinical observations regarding endocrine resistance. MicroRNAs regulate gene expression by posttranscriptional repression or degradation of mRNA, most often leading to gene silencing. MicroRNAs have been identified directly in the primary tumor, but also in the circulation of breast cancer patients. The few available studies investigating microRNA in patients suggest that seven microRNAs (miR-10a, miR-26, miR-30c, miR-126a, miR-210, miR-342 and miR-519a) play a role in tamoxifen resistance. Ingenuity Pathway Analysis (IPA) reveals that these seven microRNAs interact more readily with estrogen receptor (ER)-independent pathways than ER-related signaling pathways. Some of these pathways are targetable (e.g., PIK3CA), suggesting that microRNAs as biomarkers of endocrine resistance may have clinical value. Validation of the role of these candidate microRNAs in large prospective studies is warranted.
Keywords: biomarker; breast cancer; endocrine resistance; microRNA; tamoxifen.
Figures





Similar articles
-
Increased expression of miR-126 and miR-10a predict prolonged relapse-free time of primary oestrogen receptor-positive breast cancer following tamoxifen treatment.Eur J Cancer. 2013 Nov;49(17):3598-608. doi: 10.1016/j.ejca.2013.07.145. Epub 2013 Aug 19. Eur J Cancer. 2013. PMID: 23968733
-
MicroRNA-519a is a novel oncomir conferring tamoxifen resistance by targeting a network of tumour-suppressor genes in ER+ breast cancer.J Pathol. 2014 Aug;233(4):368-79. doi: 10.1002/path.4363. Epub 2014 Jun 2. J Pathol. 2014. PMID: 24752803 Free PMC article.
-
Cross-talk between the ER pathway and the lncRNA MAFG-AS1/miR-339-5p/ CDK2 axis promotes progression of ER+ breast cancer and confers tamoxifen resistance.Aging (Albany NY). 2020 Oct 24;12(20):20658-20683. doi: 10.18632/aging.103966. Epub 2020 Oct 24. Aging (Albany NY). 2020. PMID: 33098638 Free PMC article.
-
Identification of miRNAs as biomarkers for acquired endocrine resistance in breast cancer.Mol Cell Endocrinol. 2017 Nov 15;456:76-86. doi: 10.1016/j.mce.2017.02.004. Epub 2017 Feb 3. Mol Cell Endocrinol. 2017. PMID: 28163101 Review.
-
Tamoxifen resistance: from cell culture experiments towards novel biomarkers.Pathol Res Pract. 2015 Mar;211(3):189-97. doi: 10.1016/j.prp.2015.01.004. Epub 2015 Jan 21. Pathol Res Pract. 2015. PMID: 25666016 Review.
Cited by
-
Genome-wide analysis of therapeutic response uncovers molecular pathways governing tamoxifen resistance in ER+ breast cancer.EBioMedicine. 2020 Nov;61:103047. doi: 10.1016/j.ebiom.2020.103047. Epub 2020 Oct 21. EBioMedicine. 2020. PMID: 33099086 Free PMC article.
-
MicroRNA-139-5p modulates the growth and metastasis of malignant melanoma cells via the PI3K/AKT signaling pathway by binding to IGF1R.Cell Cycle. 2019 Dec;18(24):3513-3524. doi: 10.1080/15384101.2019.1690881. Epub 2019 Nov 14. Cell Cycle. 2019. PMID: 31724454 Free PMC article.
-
Non-Coding RNAs in Breast Cancer: Intracellular and Intercellular Communication.Noncoding RNA. 2018 Dec 12;4(4):40. doi: 10.3390/ncrna4040040. Noncoding RNA. 2018. PMID: 30545127 Free PMC article. Review.
-
Mir-30b-5p Promotes Proliferation, Migration, and Invasion of Breast Cancer Cells via Targeting ASPP2.Biomed Res Int. 2020 Apr 29;2020:7907269. doi: 10.1155/2020/7907269. eCollection 2020. Biomed Res Int. 2020. PMID: 32420372 Free PMC article. Clinical Trial.
-
MicroRNAs as a clue to overcome breast cancer treatment resistance.Cancer Metastasis Rev. 2022 Mar;41(1):77-105. doi: 10.1007/s10555-021-09992-0. Epub 2021 Sep 15. Cancer Metastasis Rev. 2022. PMID: 34524579 Free PMC article. Review.
References
-
- Haynes B.P., Viale G., Galimberti V., Rotmensz N., Gibelli B., A'Hern R., Smith I.E., Dowsett M. Expression of key estrogen-regulated genes differs substantially across the menstrual cycle in estrogen receptor-positive primary breast cancer. Breast Cancer Res. Treat. 2013;138:157–165. doi: 10.1007/s10549-013-2426-0. - DOI - PubMed
-
- Baak J.P.A., van Diest P.J., Voorhorst F.J., van der Wall E., Beex L.V.A.M., Vermorken J.B., Janssen E.A.M., Gudlaugsson E., other collaborators of the Multicenter Morphometric Mammary Carcinoma Project (MMMCP) The prognostic value of proliferation in lymph-node-negative breast cancer patients is age dependent. Eur. J. Cancer. 2007;43:527–535. - PubMed
-
- Goldhirsch A., Winer E.P., Coates A.S., Gelber R.D., Piccart-Gebhart M., Thurlimann B., Senn H.J. Personalizing the treatment of women with early breast cancer: Highlights of the St gallen international expert consensus on the primary therapy of early breast cancer 2013. Ann. Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. - DOI - PMC - PubMed
-
- Cuzick J., Dowsett M., Pineda S., Wale C., Salter J., Quinn E., Zabaglo L., Mallon E., Green A.R., Ellis I.O., et al. Prognostic value of a combined estrogen receptor, progesterone receptor, ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the genomic health recurrence score in early breast cancer. J. Clin. Oncol. 2011;29:4273–4278. doi: 10.1200/JCO.2010.31.2835. - DOI - PubMed
-
- National Comprehensive Cancer Network (NCCN) [(accessed on 4 October 2015)]. Available online: http://www.nccn.org/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

