Mining microsatellite markers from public expressed sequence tags databases for the study of threatened plants
- PMID: 26463180
- PMCID: PMC4603344
- DOI: 10.1186/s12864-015-2031-1
Mining microsatellite markers from public expressed sequence tags databases for the study of threatened plants
Abstract
Background: Simple Sequence Repeats (SSRs) are widely used in population genetic studies but their classical development is costly and time-consuming. The ever-increasing available DNA datasets generated by high-throughput techniques offer an inexpensive alternative for SSRs discovery. Expressed Sequence Tags (ESTs) have been widely used as SSR source for plants of economic relevance but their application to non-model species is still modest.
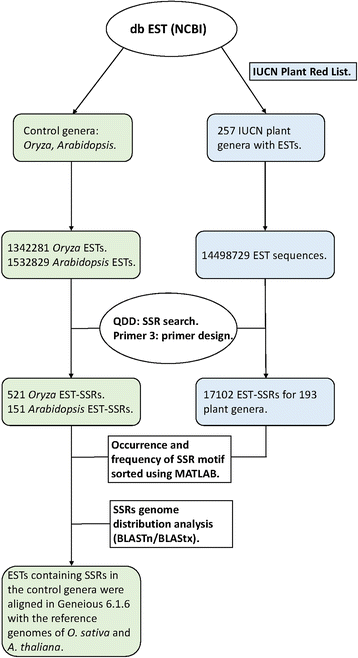
Methods: Here, we explored the use of publicly available ESTs (GenBank at the National Center for Biotechnology Information-NCBI) for SSRs development in non-model plants, focusing on genera listed by the International Union for the Conservation of Nature (IUCN). We also search two model genera with fully annotated genomes for EST-SSRs, Arabidopsis and Oryza, and used them as controls for genome distribution analyses. Overall, we downloaded 16 031 555 sequences for 258 plant genera which were mined for SSRsand their primers with the help of QDD1. Genome distribution analyses in Oryza and Arabidopsis were done by blasting the sequences with SSR against the Oryza sativa and Arabidopsis thaliana reference genomes implemented in the Basal Local Alignment Tool (BLAST) of the NCBI website. Finally, we performed an empirical test to determine the performance of our EST-SSRs in a few individuals from four species of two eudicot genera, Trifolium and Centaurea.
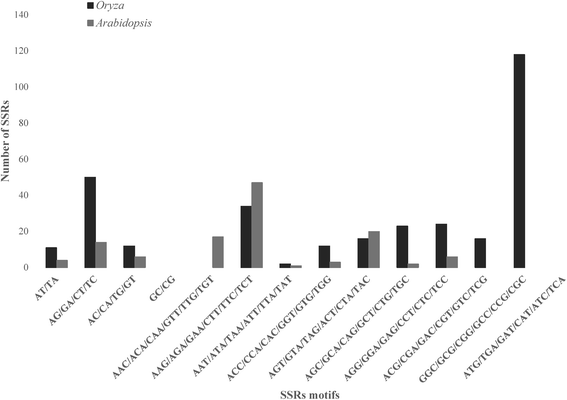
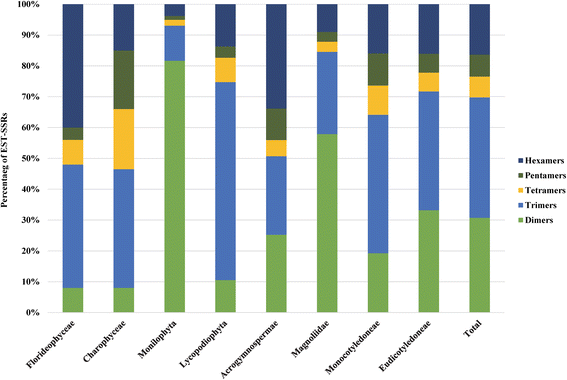
Results: We explored a total of 14 498 726 EST sequences from the dbEST database (NCBI) in 257 plant genera from the IUCN Red List. We identify a very large number (17 102) of ready-to-test EST-SSRs in most plant genera (193) at no cost. Overall, dinucleotide and trinucleotide repeats were the prevalent types but the abundance of the various types of repeat differed between taxonomic groups. Control genomes revealed that trinucleotide repeats were mostly located in coding regions while dinucleotide repeats were largely associated with untranslated regions. Our results from the empirical test revealed considerable amplification success and transferability between congenerics.
Conclusions: The present work represents the first large-scale study developing SSRs by utilizing publicly accessible EST databases in threatened plants. Here we provide a very large number of ready-to-test EST-SSR (17 102) for 193 genera. The cross-species transferability suggests that the number of possible target species would be large. Since trinucleotide repeats are abundant and mainly linked to exons they might be useful in evolutionary and conservation studies. Altogether, our study highly supports the use of EST databases as an extremely affordable and fast alternative for SSR developing in threatened plants.
Figures
Similar articles
-
An in silico mining for simple sequence repeats from expressed sequence tags of zebrafish, medaka, Fundulus, and Xiphophorus.In Silico Biol. 2005;5(5-6):439-63. In Silico Biol. 2005. PMID: 16268789
-
Mining of EST-SSR markers of Musa and their transferability studies among the members of order the Zingiberales.Appl Biochem Biotechnol. 2013 Jan;169(1):228-38. doi: 10.1007/s12010-012-9975-2. Epub 2012 Nov 21. Appl Biochem Biotechnol. 2013. PMID: 23179283
-
Transferability of cereal EST-SSR markers to ryegrass.Genome. 2009 May;52(5):431-7. doi: 10.1139/g09-019. Genome. 2009. PMID: 19448723
-
EST-SSRs as a resource for population genetic analyses.Heredity (Edinb). 2007 Aug;99(2):125-32. doi: 10.1038/sj.hdy.6801001. Epub 2007 May 23. Heredity (Edinb). 2007. PMID: 17519965 Review.
-
Genic microsatellite markers in plants: features and applications.Trends Biotechnol. 2005 Jan;23(1):48-55. doi: 10.1016/j.tibtech.2004.11.005. Trends Biotechnol. 2005. PMID: 15629858 Review.
Cited by
-
The Potential Role of Genic-SSRs in Driving Ecological Adaptation Diversity in Caragana Plants.Int J Mol Sci. 2024 Feb 8;25(4):2084. doi: 10.3390/ijms25042084. Int J Mol Sci. 2024. PMID: 38396759 Free PMC article.
-
Genetic variation and structure of endemic and endangered wild celery (Kelussia odoratissima Mozaff.) quantified using novel microsatellite markers developed by next-generation sequencing.Front Plant Sci. 2024 Apr 4;15:1301936. doi: 10.3389/fpls.2024.1301936. eCollection 2024. Front Plant Sci. 2024. PMID: 38638345 Free PMC article.
-
Complete chloroplast genomes of all six Hosta species occurring in Korea: molecular structures, comparative, and phylogenetic analyses.BMC Genomics. 2019 Nov 9;20(1):833. doi: 10.1186/s12864-019-6215-y. BMC Genomics. 2019. PMID: 31706273 Free PMC article.
-
De novo transcriptome assembly of Pueraria montana var. lobata and Neustanthus phaseoloides for the development of eSSR and SNP markers: narrowing the US origin(s) of the invasive kudzu.BMC Genomics. 2018 Jun 5;19(1):439. doi: 10.1186/s12864-018-4798-3. BMC Genomics. 2018. PMID: 29871589 Free PMC article.
-
Identification of SNP and SSR Markers in Finger Millet Using Next Generation Sequencing Technologies.PLoS One. 2016 Jul 25;11(7):e0159437. doi: 10.1371/journal.pone.0159437. eCollection 2016. PLoS One. 2016. PMID: 27454301 Free PMC article.
References
-
- Frankham R, Briscoe DA, Ballou JD. Introduction to Conservation Genetics. 2. Cambridge: Cambridge University Press; 2010.
-
- Höglund J. Evolutionary conservation genetics. Oxford: Oxford University Press; 2009.
-
- Allendorf FW, Luikart G. Conservation and the Genetics of Populations. 2. Malden: Blackwell Pub.; 2012.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

