Evolution and Structural Analyses of Glossina morsitans (Diptera; Glossinidae) Tetraspanins
- PMID: 26462947
- PMCID: PMC4592607
- DOI: 10.3390/insects5040885
Evolution and Structural Analyses of Glossina morsitans (Diptera; Glossinidae) Tetraspanins
Abstract
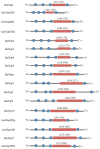
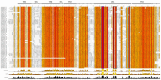
Tetraspanins are important conserved integral membrane proteins expressed in many organisms. Although there is limited knowledge about the full repertoire, evolution and structural characteristics of individual members in various organisms, data obtained so far show that tetraspanins play major roles in membrane biology, visual processing, memory, olfactory signal processing, and mechanosensory antennal inputs. Thus, these proteins are potential targets for control of insect pests. Here, we report that the genome of the tsetse fly, Glossina morsitans (Diptera: Glossinidae) encodes at least seventeen tetraspanins (GmTsps), all containing the signature features found in the tetraspanin superfamily members. Whereas six of the GmTsps have been previously reported, eleven could be classified as novel because their amino acid sequences do not map to characterized tetraspanins in the available protein data bases. We present a model of the GmTsps by using GmTsp42Ed, whose presence and expression has been recently detected by transcriptomics and proteomics analyses of G. morsitans. Phylogenetically, the identified GmTsps segregate into three major clusters. Structurally, the GmTsps are largely similar to vertebrate tetraspanins. In view of the exploitation of tetraspanins by organisms for survival, these proteins could be targeted using specific antibodies, recombinant large extracellular loop (LEL) domains, small-molecule mimetics and siRNAs as potential novel and efficacious putative targets to combat African trypanosomiasis by killing the tsetse fly vector.
Keywords: CD63; Glossina morsitans; GmTsp; LEL; Trypanosoma; modeling; phylogenetics; positive selection; tetraspanins.
Figures


Similar articles
-
Expression Levels of Odorant Receptor Genes in the Savanna Tsetse Fly, Glossina morsitans morsitans.J Med Entomol. 2018 Jun 28;55(4):855-861. doi: 10.1093/jme/tjy018. J Med Entomol. 2018. PMID: 29529232
-
Hsp70/J-protein machinery from Glossina morsitans morsitans, vector of African trypanosomiasis.PLoS One. 2017 Sep 13;12(9):e0183858. doi: 10.1371/journal.pone.0183858. eCollection 2017. PLoS One. 2017. PMID: 28902917 Free PMC article.
-
An immunoregulatory peptide from tsetse fly salivary glands of Glossina morsitans morsitans.Biochimie. 2015 Nov;118:123-8. doi: 10.1016/j.biochi.2015.09.001. Epub 2015 Sep 3. Biochimie. 2015. PMID: 26342879
-
The Emerging Role of Tetraspanins in the Proteolytic Processing of the Amyloid Precursor Protein.Front Mol Neurosci. 2016 Dec 21;9:149. doi: 10.3389/fnmol.2016.00149. eCollection 2016. Front Mol Neurosci. 2016. PMID: 28066176 Free PMC article. Review.
-
Strategies for targeting tetraspanin proteins: potential therapeutic applications in microbial infections.BioDrugs. 2009;23(6):341-59. doi: 10.2165/11315650-000000000-00000. BioDrugs. 2009. PMID: 19894777 Free PMC article. Review.
Cited by
-
Molecular characterization of tsetse's proboscis and its response to Trypanosoma congolense infection.PLoS Negl Trop Dis. 2017 Nov 20;11(11):e0006057. doi: 10.1371/journal.pntd.0006057. eCollection 2017 Nov. PLoS Negl Trop Dis. 2017. PMID: 29155830 Free PMC article.
-
A chromosome-level genome assembly of the soybean pod borer: insights into larval transcriptional response to transgenic soybean expressing the pesticidal Cry1Ac protein.BMC Genomics. 2024 Apr 9;25(1):355. doi: 10.1186/s12864-024-10216-2. BMC Genomics. 2024. PMID: 38594617 Free PMC article.
-
In silico selection of functionally important proteins from the mialome of Ornithodoros erraticus ticks and assessment of their protective efficacy as vaccine targets.Parasit Vectors. 2019 Oct 30;12(1):508. doi: 10.1186/s13071-019-3768-1. Parasit Vectors. 2019. PMID: 31666116 Free PMC article.
-
Tsetse fly tolerance to T. brucei infection: transcriptome analysis of trypanosome-associated changes in the tsetse fly salivary gland.BMC Genomics. 2016 Nov 25;17(1):971. doi: 10.1186/s12864-016-3283-0. BMC Genomics. 2016. PMID: 27884110 Free PMC article.
References
-
- Maudlin I. Transmission of African trypanosomiasis: Interactions among tsetse immune system, symbionts, and parasites. In: Harris K., editor. Advances in Disease Vector Research. 7th ed. Springer; New York, NY, USA: 1991. pp. 117–148.
-
- Vreysen M.J.B. Prospects for area-wide integrated control of tsetse flies (Diptera: Glossinidae) and trypanosomosis in sub-Saharan Africa. Rev. Soc. Entomol. Argent. 2006;65:1–21.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

