Tumor Necrosis Factor Alpha and Insulin-Like Growth Factor 1 Induced Modifications of the Gene Expression Kinetics of Differentiating Skeletal Muscle Cells
- PMID: 26447881
- PMCID: PMC4598026
- DOI: 10.1371/journal.pone.0139520
Tumor Necrosis Factor Alpha and Insulin-Like Growth Factor 1 Induced Modifications of the Gene Expression Kinetics of Differentiating Skeletal Muscle Cells
Abstract
Introduction: TNF-α levels are increased during muscle wasting and chronic muscle degeneration and regeneration processes, which are characteristic for primary muscle disorders. Pathologically increased TNF-α levels have a negative effect on muscle cell differentiation efficiency, while IGF1 can have a positive effect; therefore, we intended to elucidate the impact of TNF-α and IGF1 on gene expression during the early stages of skeletal muscle cell differentiation.
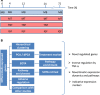
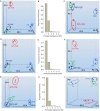
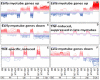
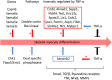
Methodology/principal findings: This study presents gene expression data of the murine skeletal muscle cells PMI28 during myogenic differentiation or differentiation with TNF-α or IGF1 exposure at 0 h, 4 h, 12 h, 24 h, and 72 h after induction. Our study detected significant coregulation of gene sets involved in myoblast differentiation or in the response to TNF-α. Gene expression data revealed a time- and treatment-dependent regulation of signaling pathways, which are prominent in myogenic differentiation. We identified enrichment of pathways, which have not been specifically linked to myoblast differentiation such as doublecortin-like kinase pathway associations as well as enrichment of specific semaphorin isoforms. Moreover to the best of our knowledge, this is the first description of a specific inverse regulation of the following genes in myoblast differentiation and response to TNF-α: Aknad1, Cmbl, Sepp1, Ndst4, Tecrl, Unc13c, Spats2l, Lix1, Csdc2, Cpa1, Parm1, Serpinb2, Aspn, Fibin, Slc40a1, Nrk, and Mybpc1. We identified a gene subset (Nfkbia, Nfkb2, Mmp9, Mef2c, Gpx, and Pgam2), which is robustly regulated by TNF-α across independent myogenic differentiation studies.
Conclusions: This is the largest dataset revealing the impact of TNF-α or IGF1 treatment on gene expression kinetics of early in vitro skeletal myoblast differentiation. We identified novel mRNAs, which have not yet been associated with skeletal muscle differentiation or response to TNF-α. Results of this study may facilitate the understanding of transcriptomic networks underlying inhibited muscle differentiation in inflammatory diseases.
Conflict of interest statement
Figures

Similar articles
-
TNF-α and IGF1 modify the microRNA signature in skeletal muscle cell differentiation.Cell Commun Signal. 2015 Jan 29;13:4. doi: 10.1186/s12964-015-0083-0. Cell Commun Signal. 2015. PMID: 25630602 Free PMC article.
-
Integrative Analysis of MicroRNA and mRNA Data Reveals an Orchestrated Function of MicroRNAs in Skeletal Myocyte Differentiation in Response to TNF-α or IGF1.PLoS One. 2015 Aug 13;10(8):e0135284. doi: 10.1371/journal.pone.0135284. eCollection 2015. PLoS One. 2015. PMID: 26270642 Free PMC article.
-
Beneficial synergistic interactions of TNF-alpha and IL-6 in C2 skeletal myoblasts--potential cross-talk with IGF system.Growth Factors. 2008 Apr;26(2):61-73. doi: 10.1080/08977190802025024. Growth Factors. 2008. PMID: 18428025
-
Sirtuin 1 regulates skeletal myoblast survival and enhances differentiation in the presence of resveratrol.Exp Physiol. 2012 Mar;97(3):400-18. doi: 10.1113/expphysiol.2011.061028. Epub 2011 Nov 28. Exp Physiol. 2012. PMID: 22125309
-
Skeletal muscle cells possess a 'memory' of acute early life TNF-α exposure: role of epigenetic adaptation.Biogerontology. 2016 Jun;17(3):603-17. doi: 10.1007/s10522-015-9604-x. Epub 2015 Sep 8. Biogerontology. 2016. PMID: 26349924 Review.
Cited by
-
Spermatogenesis associated serine rich 2 like plays a prognostic factor and therapeutic target in acute myeloid leukemia by regulating the JAK2/STAT3/STAT5 axis.J Transl Med. 2023 Feb 11;21(1):115. doi: 10.1186/s12967-023-03968-0. J Transl Med. 2023. PMID: 36774517 Free PMC article.
-
Tumor Necrosis Factor-Alpha Modulates Expression of Genes Involved in Cytokines and Chemokine Pathways in Proliferative Myoblast Cells.Cells. 2024 Jul 8;13(13):1161. doi: 10.3390/cells13131161. Cells. 2024. PMID: 38995013 Free PMC article.
-
Building Haplotype-Resolved 3D Genome Maps of Chicken Skeletal Muscle.Adv Sci (Weinh). 2024 Jun;11(24):e2305706. doi: 10.1002/advs.202305706. Epub 2024 Apr 6. Adv Sci (Weinh). 2024. PMID: 38582509 Free PMC article.
-
Discovery of exercise-related genes and pathway analysis based on comparative genomes of Mongolian originated Abaga and Wushen horse.Open Life Sci. 2022 Sep 26;17(1):1269-1281. doi: 10.1515/biol-2022-0487. eCollection 2022. Open Life Sci. 2022. PMID: 36249530 Free PMC article.
-
lncRNA DLEU2 acts as a miR-181a sponge to regulate SEPP1 and inhibit skeletal muscle differentiation and regeneration.Aging (Albany NY). 2020 Nov 18;12(23):24033-24056. doi: 10.18632/aging.104095. Epub 2020 Nov 18. Aging (Albany NY). 2020. PMID: 33221762 Free PMC article.
References
-
- Droguett R, Cabello-Verrugio C, Riquelme C, Brandan E. Extracellular proteoglycans modify TGF-beta bio-availability attenuating its signaling during skeletal muscle differentiation. Matrix Biol. 2006; 25: 332–341. - PubMed
-
- Foulstone EJ, Huser C, Crown AL, Holly JM, Stewart CE. Differential signalling mechanisms predisposing primary human skeletal muscle cells to altered proliferation and differentiation: roles of IGF-I and TNFalpha. Exp. Cell Res. 2014; 294: 223–235. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

